Figure 1.
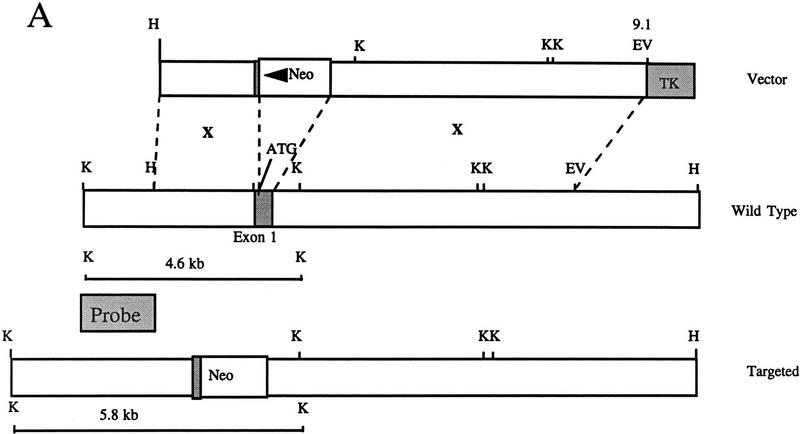
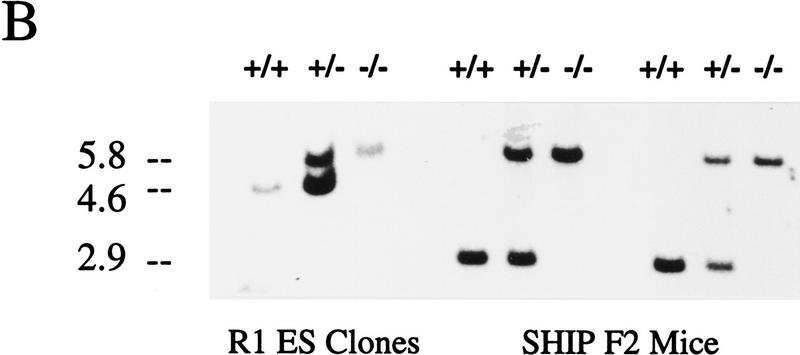
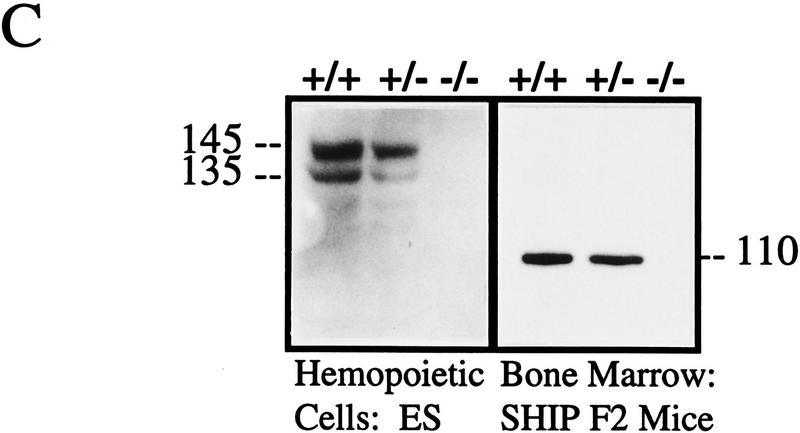
Targeted disruption of the murine SHIP gene. (A) Partial restriction map of the wild-type 129 SHIP gene (Wild type), the tk-containing targeting vector that replaces the coding sequence of the first exon with the Neo-resistance gene in the antisense orientation (Vector), and the organization of the targeted 129 allele (Targeted). (Dashed lines) Area of homology between the vector and the endogenous gene. The 2.0-kb genomic probe used for screening is indicated along with expected sizes of the wild-type and targeted KpnI fragments. (B) Genomic Southern blot analysis using a 2-kb KpnI–HindIII genomic probe. The nontargeted SHIP allele is visualized as a 4.6-kb band in the wild-type ES cells and a 2.9-kb C57Bl/6 band in the mice (because of a polymorphism at this locus), whereas the SHIP−/− cells and mice contain only the targeted 5.8-kb band. (C) Western blot analysis. R1 SHIP wild-type (+/+), heterozygous (+/−), and null (−/−) ES cells were differentiated in vitro as described previously (Helgason et al. 1996). Hemopoietic cells were derived from suspension cultures of day 10 embryoid bodies, and total cell lysates were probed with an antibody against the carboxy-terminal region spanning the two NPxY sequences of SHIP. The predominant 145- and 135-kD forms are indicated at left. Total cell lysates were also prepared from freshly isolated bone marrow of representative F2 mice and blots were probed with an antibody against the amino terminal SH2 domain. The predominant 110-kD protein is indicated at right.