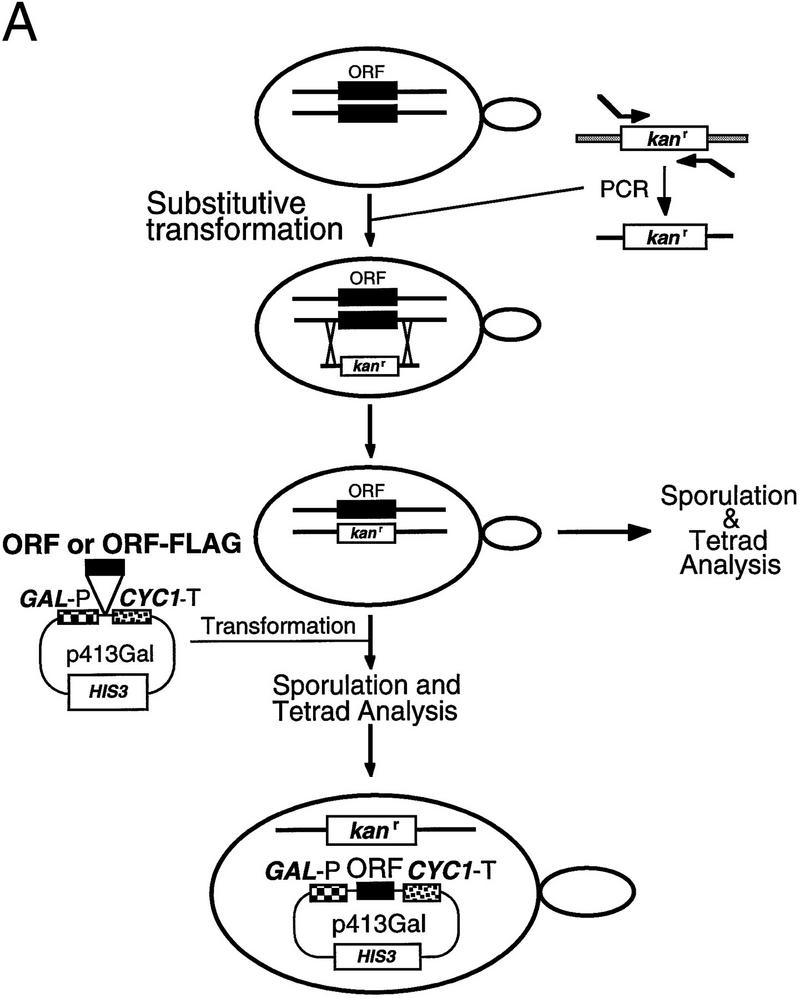
Figure 2.
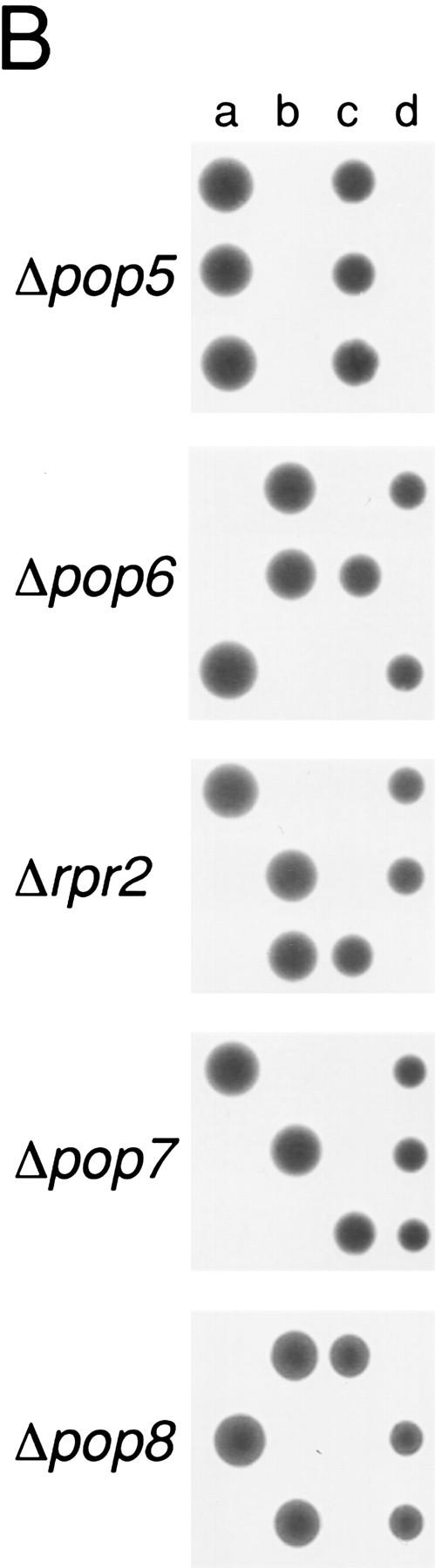
All of the newly identified protein subunit candidates are encoded by essential genes. Four of the proteins that copurify with RNase P activity (Pop1p, Rpp1p, Pop4p, and Pop3p) had been identified previously as candidate RNase P subunits, and their genes had been shown to be essential. The genes for the remaining five subunits (Pop5p, Pop6p, Rpr2p, Pop7p, and Pop8p) were tested to determine whether they were essential for viability and if depletion of their gene products affected RNase P or RNase MRP function. (A) The experimental design for deleting the coding regions and substituting a gene under the regulation of a GAL1 promoter is shown schematically. Homologous recombination was used to replace one copy of each coding region in diploids with a gene for kanamycin resistance. The resulting heterodiploids were first sporulated and tested for spore viability. B shows that all tetrads segregated as two viable and two nonviable spores, the result expected for essential genes. The diagram also shows a schematic for creation of a haploid strain in which the genomic copy of the gene of interest is disrupted, and production of the gene product from a plasmid is placed under Gal control for the depletion studies shown in Fig. 3. These recombinant gene products were also all made in two versions, both of which conferred viability. One version, used for the RNA blot analyses in Fig. 3, contains only the coding region for the gene. A second version, used in the immunoprecipitation experiments in Fig. 4, also contains the 8-amino-acid Flag epitope at the carboxyl terminus.