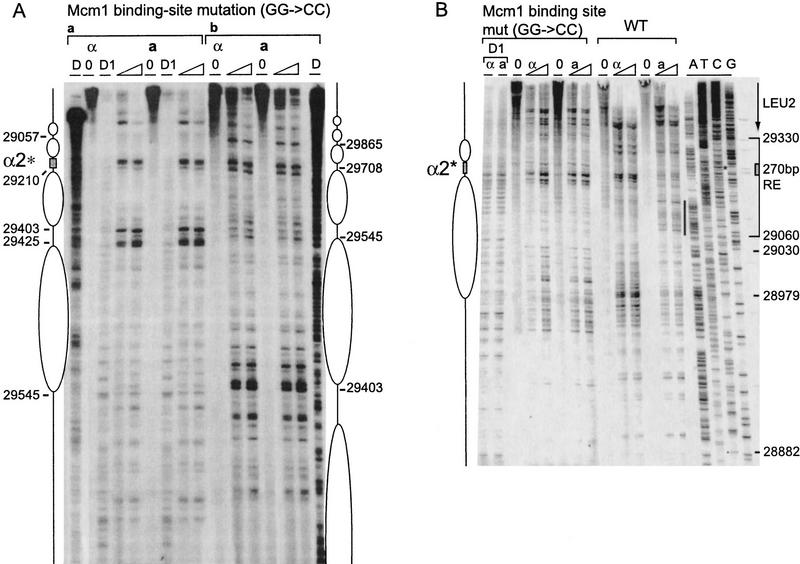
Figure 4.
Effect of Mcm1 binding site mutation on RE chromatin structure. (A) Chromatin structure of the S. cerevisiae 753-bp RE fragment containing a 2-bp mutation (GG to CC at positions 29212–29213) in the Mcm1 binding site of the α2 operator mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer b297 (a) and a292 (b). Mata and Matα cells are as indicated (CWU57, CWU94). Extensions of undigested chromatin (0) and chromatin digested with two levels of micrococcal nuclease are presented. (D) Protein-free DNA digests. The mutated Mcm1–Matα2 binding site consensus sequence (α2*) is represented by a shaded box. (B) Chromatin structure of the S. cerevisiae 270-bp minimal RE fragment containing the wild-type α2 consensus sequence (29194–29225) or a point mutation (GG-CC at positions 29212–29213) in the Mcm1 binding site mapped by primer extension analysis of micrococcal nuclease cleavage sites with primer a288. MATa and MATα cells are as indicated (CWU81 and CWU101 for wild-type 270 bp, CWU137 and CWU143 for Mcm1 binding site mutation, respectively). Extensions of undigested chromatin (0) and two levels of micrococcal nuclease-digested chromatin are presented. (D1) DNase I-digested chromatin. (*) The position of the mutation in the Mcm1 binding sequence. The limits of the 270-bp fragment are indicated by a bracket. The ellipse corresponds to the inferred position of a nucleosome. The vertical black stripe emphasizes the area of increased nuclease cleavage in wild-type MATa vs. MATα cells. The 3′ end of the LEU2 transcript is indicated by an arrow. The positions of the sequences were determined by four dideoxy sequencing reactions with the same primer (lanes A, T, G, and C).