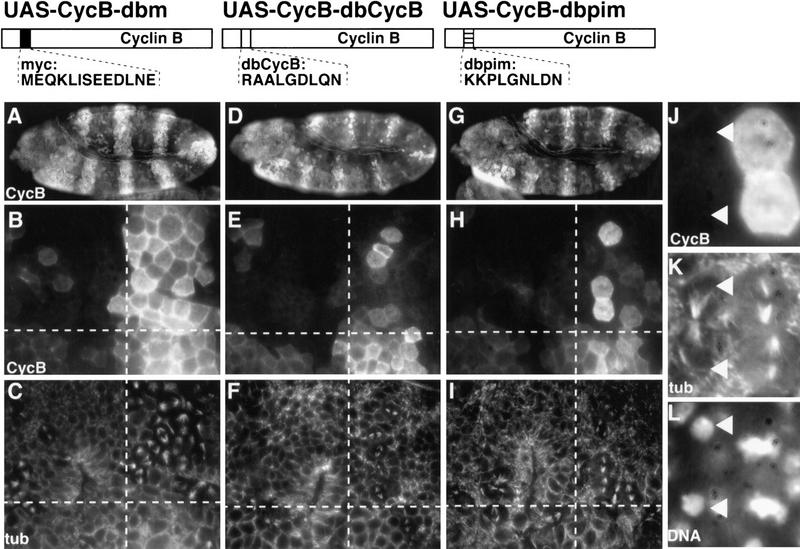
Figure 2.
The PIM destruction box variant can replace the Cyclin B destruction box. prd–GAL4, which directs UAS target gene expression in alternating segments starting before mitosis 15 of Drosophila embryogenesis, was used to express either Cyclin B with the myc epitope in place of the destruction box (UAS–CycB–dbm; A–C), or Cyclin B with the endogenous destruction box (UAS–CycB–dbCycB; D–F), or Cyclin B with the PIM destruction box (UAS–CycB–dbpim; G–L). Embryos (A,D,G) were fixed during the stage of mitosis 15 and double-labeled with antibodies against Cyclin B (CycB; A,B,D,E,G,H,J), tubulin (tub; C,F,I,K) and a DNA stain (DNA; L). Higher magnification views of the embryonic epidermis (B,C,E,F,H,I) are shown with the regions of UAS target gene expression to the right of the dashed vertical lines. UAS target gene expression is absent from the regions on the left of the dashed vertical line. These regions express only endogenous Cyclin B and serve as internal control for progression through mitosis 15. Progression through mitosis 15 is accompanied by degradation of Cyclin B protein when carrying a functional degradation box and occurs in a segmentally repeated pattern (Foe et al. 1993) first in the dorsal epidermis (above the horizontal dashed line) and only later in the ventral epidermis (below the horizontal dashed line). At the stage shown, mitosis 15 is largely completed in the dorsal epidermis and just starting in the ventral epidermis. Nondegradable Cyclin B with the myc epitope in place of the destruction box blocks exit from mitosis and results in an enrichment of mitotic figures (C, upper right) in cells that are labeled by anti-Cyclin B (B, upper right). In contrast, Cyclin B with the PIM motif in place of the destruction box does not block exit from mitosis and is degraded during late mitosis as illustrated in the regions shown at even higher magnification (J–L). Arrowheads mark a telophase cell that is not labeled with anti-Cyclin B (J) while the neighboring metaphase cells (right) are strongly labeled. The structure of the different UAS-transgenes is schematically illustrated above the panels with the corresponding results.