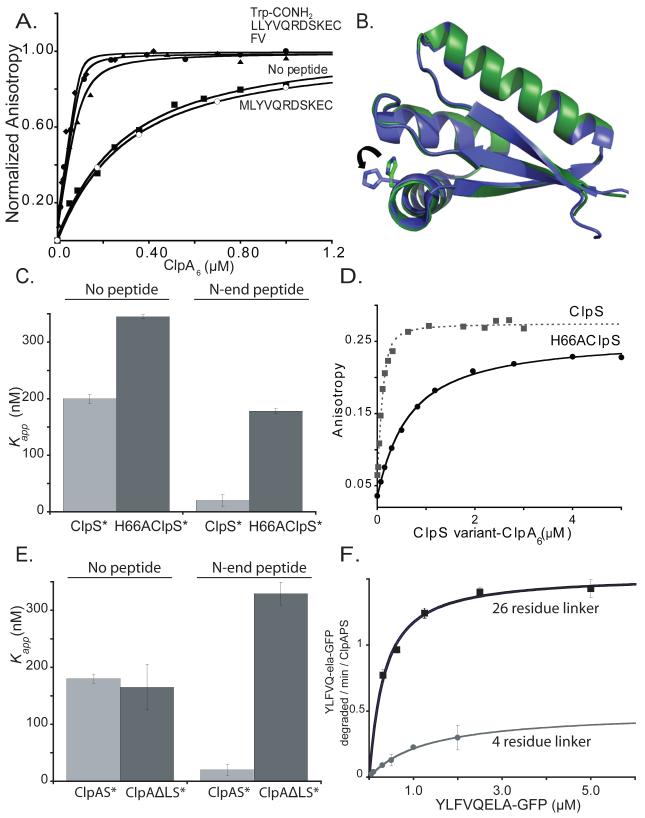
Figure 3. ClpS binds ClpA6 more tightly in the presence of N-end-rule peptides.
A) As assayed by anisotropy, ClpA6 bound 200 nM fluorescent ClpS*F tightly in the presence 20 μM Trp-conh2, LLYVQRDSKEC, or FV N-end-rule peptides (Kapp < 20 nM) and more weakly in the absence of peptide or with 20 μM MLYVQRDSKEC peptide (Kapp ~ 180 nM).
B) The H66 residue of ClpS is one of the side chains involved in the formation of one of the three hydrogen bonds that the adaptor forms with the α-amino group of the N-end degron. Overlay of the apo (green, PDB 3O1F) and the peptide-bound (blue, 2W9R) crystal structures of ClpS reveal no major global changes occur upon peptide binding. The most substantial change is the rotation of the H66 side chain, which appear to need to move away from the pocket in the apo form to accommodate the N-degron in the peptide binding site.
C) ClpA6 bound ClpS*F (KD = 200 ± 6 nM) and H66AClpS*F (KD = 345 ± 3 nM) with similar affinities. Addition of 20 μM LLYVQRDSKEC N-end-rule peptide enhanced ClpA6 affinity for ClpS*F substantially (Kapp = 20 ± 10 nM) but increased affinity for H66AClpS*F only modestly (Kapp = 178 ± 4 nM).
D) H66AClpS-ClpA6 complex binds more weakly to an N-end-rule fluorescent peptide (LLYVQRDSKEC-fl) when compared to ClpS-ClpA6 (Kapp = 560 nM vs Kapp = 42 ± 6 nM for wild type).
E) An N-end-rule peptide (LLYVQRDSKEC; 20 μM) enhanced binding of ClpS*F to ClpA6 but not to ΔLClpA6, which has shorter linker between the N and D1 domains (Cranz-Mileva et al., 2008).
F) Michaelis-Menten plots showed that substituting ΔLClpA6 (4-residue linker) for ClpA6 (26-residue linker) decreased KM and Vmax for ClpAPS degradation (100 nM ClpA6 or ΔLClpA6; 200 nm ClpP14; 600 nm ClpS) of the N-end-rule substrate ylfvqela-GFP.