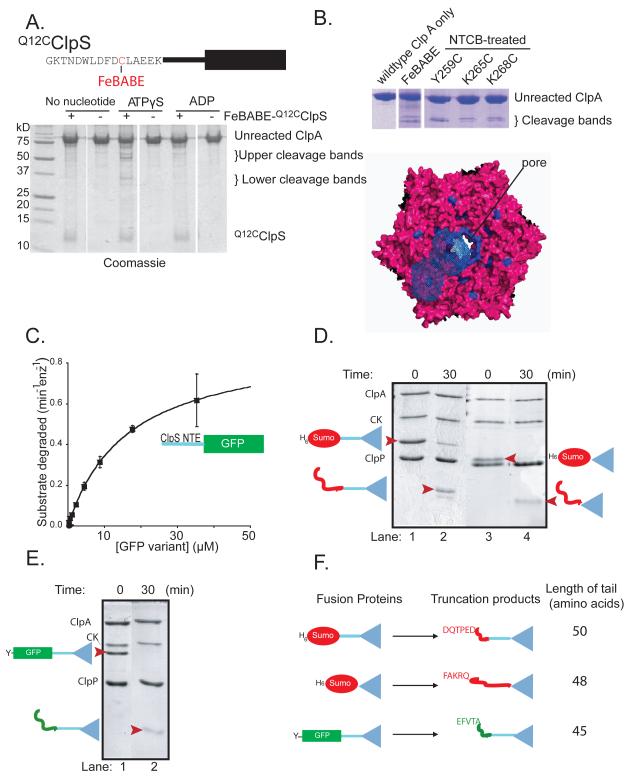
Figure 5. The ClpS NTE contacts ClpA near the axial pore.
A) Top: FeBABE was attached to residue 12 of Q12CClpS variant for cleavage studies. Bottom: As assayed by SDS-PAGE, cleavage of ClpA required FeBABE-modified Q12CClpS and ATPγS.
B) ClpA residues 259-268 are highlighted in blue in a top view of a model of the hexameric D1 ring (Guo et al., 2002). In one ClpA subunit, blue-wire shading shows regions within 12 Å of residues 259-268, which represents the approximate reach of the tethered FeBABE.
C) A substrate consisting of residues 2-26 of ClpS fused to GFP was efficiently degraded by ClpAP, as shown by Michaelis-Menten analysis (KM = 16.4 μM; Vmax = 0.62 min−1 enz−1).
D) Assays monitored by SDS-PAGE showed that ClpAP only partially degraded the H6-Sumo-ClpS and H6-Sumo-ClpScore fusion proteins, resulting in truncated products of a lower molecular weight (marked by red arrowheads in lanes 2 & 4).
E) ClpAP partially degraded the ylfvqela-GFP-ClpS fusion protein, resulting in a lower molecular weight truncation product (marked by a red arrowhead in lane 2).
F) Depiction of the ClpS fusion proteins used to test degradation by ClpAP (left) and the corresponding truncation products produced by degradation (right). N-terminal sequencing of the truncation products revealed that the new N-termini corresponded to an internal sequence in the protein fused to ClpS (either Sumo or GFP). The truncation products consisted of the ClpS core and an additional N-terminal tail of 45-50 amino acids.