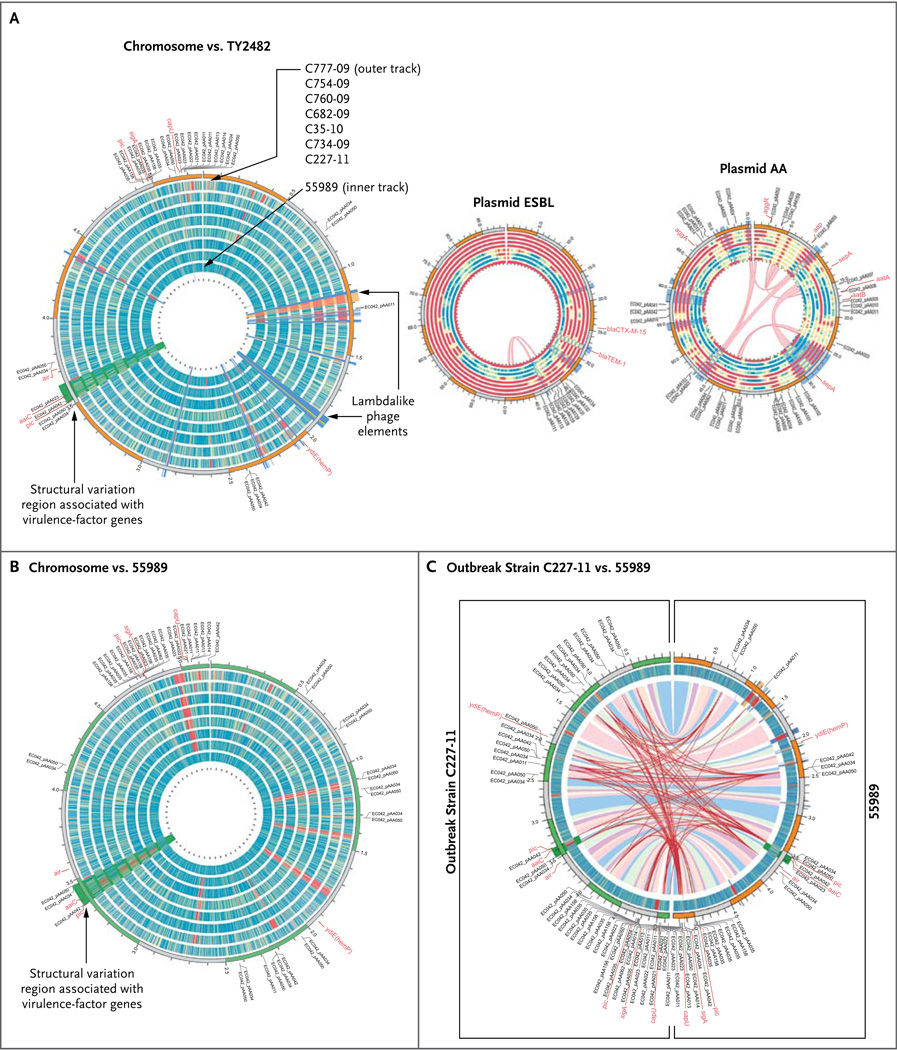
Figure 1. Comparison of Genomes of Eight Enteroaggregative Escherichia coli O104:H4 Isolates.
In Panel A, the eight circular bands represent the tracks for the different enteroaggregative E. coli (EAEC) O104:H4 isolates sequenced in our study (from inner to outer tracks: 55989, C227-11, C734-09, C35-10, C682-09, C760-09, C754-09, and C777-09). These different bands represent coverage plots mapped against the reference genome TY2482, with blue or green indicating a high degree of coverage, yellow moderate coverage, and orange or red little or no coverage. The C227-11 coverage plot indicates near-complete sequence identity with the TY2482 genome. The annotations on the outside of the chromosome plot highlight positions of known virulence-factor genes and genes previously characterized on the AA plasmid in the E. coli O42 strain. The band ordering and color coding for the plasmids are the same as those described for the chromosome plot. The thin red lines in the pAA plot show internal repeats between 55989 and C227-11 that highlight shuffling of these elements around the plasmid sequence. In Panel B, the reference genome is 55989. Panel C shows a comparison between the genome of the outbreak strain C227-11 (left semicircle) and the 55989 genome (right semicircle), mapped against the 55989 genome and the genome of the outbreak strain TY2482, respectively. The colored ribbons inside the innermost track (blue, pink, and purple) represent homologous mappings between the two genomes, C227-11 and 55989, indicating a high degree of similarity between these genomes. The red lines show the chromosomal positioning of repeat elements, such as insertion sequences and other mobile elements, which reveal more heterogeneity between the genomes than is evident from mapping regions of homology.