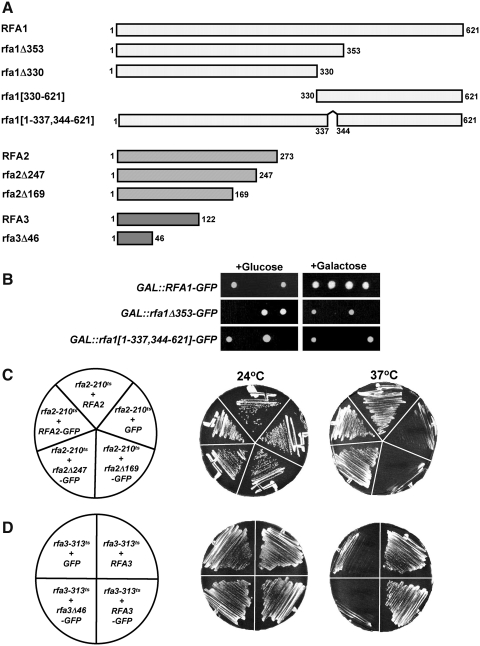
FIG. 1.
Complementation analysis of Rfa-GFP fusions. (A) Full-length and truncated Rfa1-, Rfa2-, and Rfa3-GFP chimeras were generated with GFP fused to the C-terminus of each. The numbers shown indicate the amino acid present at the N- and C-termini of each Rfa fusion encoded in each construct. (B) Diploid yeast heterozygous for an rfa1Δ deletion were transformed with full-length RFA1-GFP, rfa1Δ353-GFP, or rfa1[1–337,344–621]-GFP expressed under the control of the GAL1 promoter. Diploid transformants were sporulated and resulting tetrads dissected on selective media containing glucose (left) or galactose (right) as a carbon source. The growth of colonies from a single representative tetrad from each plate is depicted. (C) Haploid yeast containing the rfa2-210 temperature-sensitive mutation (Maniar et al., 1997) were transformed with plasmids expressing wild-type RFA2 (RFA2), empty vector (GFP), full-length RFA2 fused to GFP (RFA2-GFP), and two C-terminal rfa2 truncations fused with GFP (rfa2Δ169-GFP and rfa2Δ247-GFP). Transformants were streaked on selective media at permissive and restrictive temperatures assayed for growth. Images were taken 3 and 2 days after streaking to 24°C and 37°C, respectively. (D) Haploid cells containing rfa3-313ts were transformed with plasmids expressing wild-type RFA3 (RFA3), GFP alone (GFP), and full-length (RFA3-GFP) and truncated (rfa3Δ46-GFP) alleles of RFA3. Complementation was determined by assaying for growth at 24°C and 37°C after 48–72 h.