Figure 4.
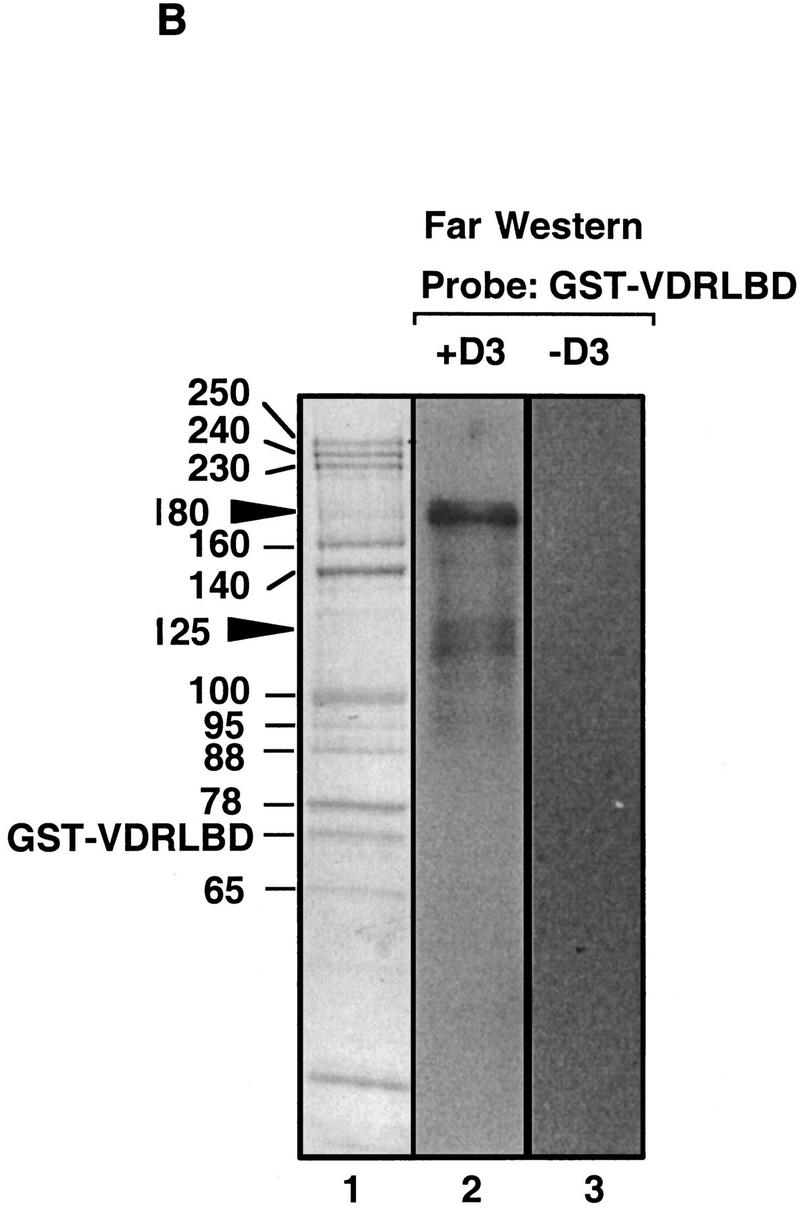
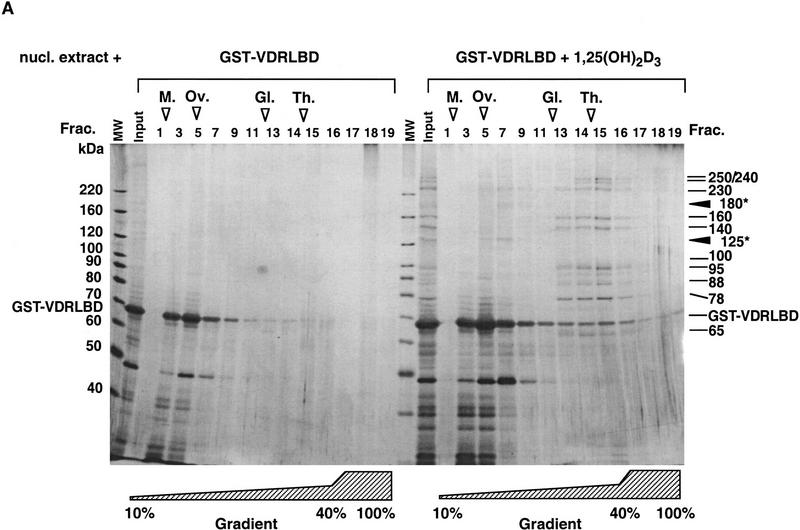
DRIPs bind to VDR LBD as a complex. (A) Glycerol gradient analysis: Nuclear extracts were passed over immobilized GST–VDR LBD in the absence or presence of 1,25(OH)2D3; receptors were then eluted from glutathione beads with reduced glutathione, and applied to 10%–40% glycerol gradients. (Left) SDS-PAGE analysis of glycerol gradient fractions of GST–VDR LBD in the absence of 1,25(OH)2D3; (right) fractions of GST–VDR LBD in the presence of 1,25(OH)2D3. Gradient fractions are from 1–17, with a 100% cushion at the bottom of the gradient (fractions 18 and 19). The position of marker proteins of known molecular masses as they sedimented through the gradient are shown above the fraction numbers: Myoglobin, 17 kD; ovalbumin, 44 kD; gamma globulin, 158 kD; thyroglobulin, 670 kD. Note that uncomplexed GST–VDR LBD, with an apparent molecular mass of 70 kD, sedimented between fractions 5–7 in both gradients. Asterisks denote two proteins of 180 and 125 kD, present in the input that cosediment with GST–VDR LBD (fractions 5–7). (B) Far Western blot. (Lane 1) Coomassie stain of the 1,25(OH)2D3-dependent DRIP complex. (Lanes 2,3) Far Western blot of the DRIP protein eluate (10 μg total protein) probed with 32P-labeled GST–VDR LBD in the presence (lane 2) or absence (lane 3) of 10−7 m 1,25(OH)2D3. Note that the exposure time of the blot probed with unliganded VDR LBD was four times longer than the same blot probed with liganded VDR LBD (4 days vs. 1 day). The solid arrowheads denote the detectable protein bands on the blot, running with apparent molecular mass of 180 and 125/130 kD.