Abstract
Phylogeographic studies provide critical insight into the evolutionary histories of model organisms; yet, to date, range-wide data are lacking for the rough periwinkle Littorina saxatilis, a classic example of marine sympatric speciation. Here, we use mitochondrial DNA (mtDNA) sequence data to demonstrate that L. saxatilis is not monophyletic for this marker, but is composed of two distinct mtDNA lineages (I and II) that are shared with sister species Littorina arcana and Littorina compressa. Bayesian coalescent dating and phylogeographic patterns indicate that both L. saxatilis lineages originated in the eastern North Atlantic, around the British Isles, at approximately 0.64 Ma. Both lineages are now distributed broadly across the eastern, central and western North Atlantic, and show strong phylogeographic structure among regions. The Iberian Peninsula is genetically distinct, suggesting prolonged isolation from northeastern North Atlantic populations. Western North Atlantic populations of L. saxatilis lineages I and II predate the last glacial maximum and have been isolated from eastern North Atlantic populations since that time. This identification of two distinct, broadly distributed mtDNA lineages further complicates observed patterns of repeated incipient ecological speciation in L. saxatilis, because the sympatric origins of distinct ecotype pairs on eastern North Atlantic shores may be confounded by admixture of divergent lineages.
Keywords: speciation, phylogeography, glacial refugia, trans-Arctic interchange, cryptic diversity, mitochondrial DNA
1. Introduction
The evolutionary histories of North Atlantic benthic marine species have been shaped by the trans-Arctic interchange (TAI) and subsequent, repeated glaciations throughout the Pleistocene [1–3]. Following the opening of a trans-Arctic migration route around 5.5 Ma, the TAI (5.5–2.4 Ma) introduced numerous Pacific species to novel selective regimes in the North Atlantic [1,4,5]. Ensuing glacial cycles drove oscillations in sea level and habitat availability, which contracted populations into limited refugia during glacial maxima, and yielded repeated range fluctuations, population subdivision and regional extinctions on both sides of the North Atlantic [2–4].
The last glacial maximum (LGM), at approximately 18 Ka, has received significant attention because phylogenetic data demonstrate that its effects differed markedly between populations of marine organisms on eastern and western North Atlantic shores [3,4]. In the eastern North Atlantic, there is evidence for multiple southern glacial refugia, which buffered extinctions and allowed for northward recolonization events during interglacial periods [6–9]. In the western North Atlantic, regional extinctions were common among rocky shore species, because most of the suitable habitat was covered by the Laurentide ice sheet [2,8,10], although some species were able to survive in southern [2,11] or northern refugia [3,7,11–14]. As a result, many of the current western North Atlantic rocky shore fauna (including the intertidal snails Littorina obtusata and Nucella lapillus) recolonized the region following the LGM, whereas other species (such as Semibalanus balanoides and Mytilus edulis) appear to have persisted in western North Atlantic refugia [2–4,8,10].
Amid the extensive climatic variation of the past 5.5 Myr, Littorina (subgenus Neritrema) snails underwent a relatively high incidence of speciation, giving rise to five North Atlantic species, including three rough periwinkles: egg-laying Littorina compressa and Littorina arcana, and the brooding species Littorina saxatilis [1,15,16]. The brooder L. saxatilis has the broadest range, spanning the North Atlantic from northern Africa to the mid-Atlantic coast of North America, while both egg-laying species have narrow distributions restricted to the northeastern North Atlantic coasts of Europe (e.g. France, the British Isles and Norway [15]).
Littorina saxatilis is also notable for its wide range of morphologies [15], especially in eastern North Atlantic regions (e.g. Sweden, Spain and Britain), where it exhibits extreme local (i.e. on the scale of metres) habitat-specific morphological differentiation driven primarily by variation in crab predation and wave exposure [15,17,18]. In each location, pairs of divergent ecotypes display distinct morphologies (E & S, Sweden [19]; RB & SU, Spain [20]; H & M, Britain [21]), partial reduction of gene flow [22–25] and evidence of reproductive isolation [21,26,27], providing support for multiple independent divergence events and suggesting L. saxatilis as a model of incipient sympatric speciation [17,18,26,28]. Yet, despite its importance as one of the few marine examples of incipient sympatric speciation [29], the necessity of lineage-wide phylogenetic context for model systems of sympatric speciation [30,31] and the established role of historical allopatric divergence in North Atlantic marine species [3,4], little is known about the evolutionary history of L. saxatilis across its trans-Atlantic range.
Although extensive localized genetic studies of rough periwinkles in the eastern North Atlantic have been conducted, western North Atlantic populations remain largely unexamined, and no comprehensive evaluation of range-wide genetic variation within L. saxatilis exists. Here, we use mitochondrial DNA (mtDNA) sequence data and Bayesian phylogenetic analysis to reconstruct the evolutionary relationships among L. compressa, L. arcana and a range-wide sampling of L. saxatilis. Using an mtDNA gene tree, we ask whether these species are genetically distinct (i.e. reciprocally monophyletic), date the origins of distinct mtDNA lineages and define the phylogeographic structure of L. saxatilis populations across the North Atlantic.
2. Material and methods
(a). Sampling and molecular analysis
We sampled 405 individuals from 32 sites (electronic supplementary material, table S1) across the range of L. saxatilis (western, central and eastern North Atlantic). Additionally, we collected 33 L. arcana and 20 L. compressa from the northeastern North Atlantic. To confirm specimen identity in areas of sympatry, we used only females with a clearly defined brood pouch (L. saxatilis) or jelly gland (L. arcana and L. compressa). Littorina compressa was further distinguished by distinctive shell grooves [15]. Snails were dissected live, frozen at −80°C or preserved in 100 per cent ethanol prior to extraction. Whole genomic DNA was extracted from head/foot tissue using hexadecyltrimethylammonium bromide extraction buffer and a standard phenol/chloroform extraction protocol [32]. We designed two new primers (LsaxMt_F, 5′-CTG ATG CCG CAA AAC TTC TT-3′ and LsaxMt_R, 5′-GTC AAC TGC AAA GCC TCC TC-3′) from published sequences [33,34] and amplified a 1832 bp region of the mitochondrial genome, including NADH1, tRNApro, NADH6 and partial cytochrome b. PCR amplifications were obtained using Phire Hot Start DNA Polymerase (Finnzymes) and a thermal profile of 30 s at 98°C, followed by 30 cycles of 98°C for 5 s, 50°C for 5 s and 72°C for 1 min and 15 s. PCR products were cleaned and sequenced in both directions using the PCR primers on ABI 3700 capillary sequencers by Agencourt Bioscience (Beverly, MA, USA). A 633 bp sequence was obtained for NADH1 using the forward primer (GenBank accession nos JF501729–JF501840). From the reverse primer, 521 bp were obtained, including 61 bp of NADH6 and 451 bp of cytochrome b (GenBank accession nos JF501841–JF501952). We also included the 48 published sequences of Quesada et al. [34] (EMBL-Bank accession nos AM500945–AM500965) for a total of 453 L. saxatilis samples (electronic supplementary material, table S1).
(b). DNA sequence analyses
We aligned and manually edited sequences using Sequencher v. 4.8 (Gene Codes Corporation). Mitochondrial DNA sequence variation was analysed using DnaSP v. 5.10 [35] by species, geographical region (Iberian Peninsula, and eastern, central and western North Atlantic) and population. Calculations for lineage-specific groupings were made based on the results of the phylogenetic analyses described below. Our estimates also include standard tests for deviations from neutrality, including Tajima's D, Fu and Li's D* and Fu and Li's F* [36,37]. A maximum parsimony network was constructed with TCS v. 1.21 [38].
(c). Phylogenetic analyses and divergence estimates
Phylogenetic analyses were conducted using a Bayesian approach in BEAST v. 1.5.3 [39]. The best-fit model of sequence evolution was estimated as GTR + G + I using AIC criterion generated in MrModeltest v. 2.3 [40]. This model was implemented in BEAST under strict clock and relaxed uncorrelated lognormal frameworks [41], using the assumption of a constant coalescent population size parameter. Bayes factor tests were used to evaluate differences in outcome under alternative demographic models and revealed that our data fit the molecular clock assumption [42]. By including representative sequences from Littorina fabalis and L. obtusata [34] (EMBL-Bank accession nos AM500966–AM500967) as outgroups and applying a prior probability distribution to the root of this tree, we were able to estimate the times of divergence (i.e. the time to most recent common ancestor, TMRCA) for lineages of interest. The analyses were calibrated with a combination of fossil evidence and published times of divergence among members of the North Atlantic Neritrema. With a lognormal prior, we fixed the minimum time of divergence at 2 Ma, as this is the earliest known occurrence of distinct flat periwinkles (L. fabalis) and rough periwinkles (Littorina islandica, the extinct proposed ancestor of the L. saxatilis group) in the fossil record [15]. We used the previously published mean estimate for divergence among all North Atlantic Neritrema (2.83 Ma [43]) as the mean root age, and adjusted the standard deviation so that the upper bound of the 95 per cent high probability density (HPD) approximated the opening of the Bering Strait (approx. 5.5 Ma). We also examined lineages for signatures of population size fluctuations using the GRMF Skyride option in BEAST [44]. Applying a normally distributed clock rate prior, equivalent to that estimated for the full tree, these analyses provided more conservative estimates of TMRCA than the constant population size models. All analyses were run with chain lengths of 30 million, sampling every 3000, and model performance was assessed in Tracer v. 1.5 [45]. We constructed maximum clade credibility phylogenies in TreeAnnotator v. 1.5.3 [39].
3. Results
(a). Genetic diversity
Analysis of the mtDNA fragment produced an alignment of 1154 bp with 125 variable sites and 113 unique haplotypes. Across the North Atlantic, L. saxatilis (n = 453 individuals) haplotype diversity is high (mean + s.e. = 0.94 + 0.01), with 95 unique haplotypes (electronic supplementary material, table S2). Populations on western (n = 197) and eastern (n = 107) North Atlantic shores have equal haplotype diversities (both 0.85 + 0.02), although a greater number of unique haplotypes were recovered from the western North Atlantic (h = 45 versus 25). The Iberian Peninsula (n = 52) shows the highest regional haplotype diversity (0.89 + 0.02), with 14 unique haplotypes. Haplotype diversity in the central North Atlantic (n = 97) is lowest, at 0.79 + 0.04 (h = 18). Across the eastern North Atlantic, L. arcana (n = 33, h = 12) and L. compressa (n = 20, h = 9) show comparable haplotype diversities of 0.73 + 0.08 and 0.79 + 0.09, respectively.
(b). Gene tree and divergence estimates
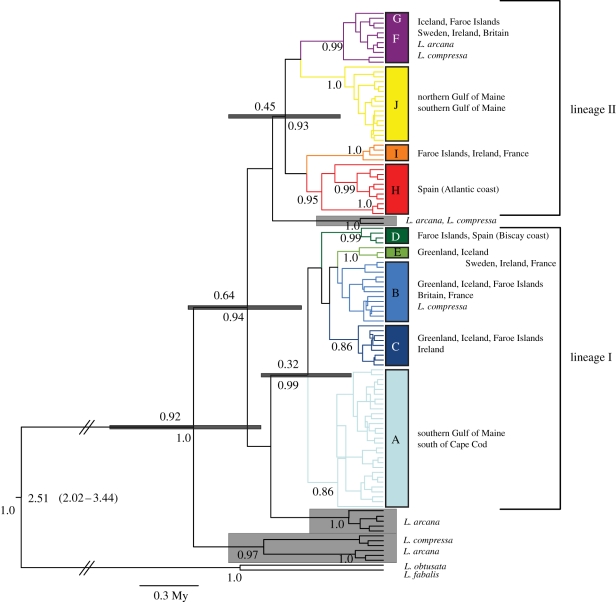
TMRCA estimates place the origin of the North Atlantic Neritrema at 2.52 Ma (95% HPD 2.01–3.50; table 1). The three rough periwinkles—L. compressa, L. arcana and L. saxatilis—form a well-supported monophyletic clade (figure 1), with a TMRCA of 0.92 Ma (0.51–1.44). None of the three species is strictly monophyletic; rather, each is made up of multiple divergent mtDNA lineages, most of which are shared among species. The majority of L. compressa and one L. arcana individual possess a basal mtDNA lineage that originated at approximately 0.92 Ma. All L. saxatilis, the majority of L. arcana and the remaining L. compressa mtDNA haplotypes form a single well-supported clade (posterior probability: 0.94) with an age of 0.64 Ma (0.35–1.00). This clade is composed of four well-supported lineages, with L. saxatilis falling into two distinct, strongly supported mtDNA lineages, I and II, which originated approximately 0.64 Ma. Lineage I also includes L. compressa, whereas lineage II contains both L. arcana and L. compressa. Two additional mtDNA lineages, one composed of L. arcana only and the other including L. compressa and L. arcana, also fall within this clade. These mtDNA lineages originated around the same time as lineages I and II (approx. 0.64 Ma); however, their phylogenetic relationships to lineages I and II remain unresolved. All three species share one identical lineage II haplotype (F), while a second is shared between L. saxatilis and L. arcana (figure 2a).
Table 1.
Estimated node ages and time to most recent common ancestor (TMRCA) in millions of years (mean, median, 95% high probability density).
| TMRCA (Ma) |
node age (Ma) |
|||||
|---|---|---|---|---|---|---|
| mean | median | 95% HPD | mean | median | 95% HPD | |
| Neritrema | 2.52 | 2.36 | 2.01–3.50 | 2.51 | 2.35 | 2.02–3.44 |
| rough periwinkles | 0.92 | 0.87 | 0.51–1.44 | 0.92 | 0.87 | 0.50–1.41 |
| L. compressa | 0.92 | 0.87 | 0.50–1.44 | 0.92 | 0.87 | 0.50–1.41 |
| L. arcana | 0.92 | 0.87 | 0.50–1.44 | 0.92 | 0.87 | 0.50–1.41 |
| L. saxatilis | 0.62 | 0.59 | 0.32–0.98 | 0.64 | 0.61 | 0.35–1.00 |
| lineage I | 0.32 | 0.30 | 0.15–0.53 | 0.32 | 0.30 | 0.15–0.53 |
| lineage II | 0.45 | 0.43 | 0.23–0.73 | 0.45 | 0.43 | 0.23–0.71 |
| lineage I western | 0.19 | 0.17 | 0.07–0.35 | 0.17 | 0.15 | 0.07–0.28 |
| lineage II western | 0.14 | 0.13 | 0.05–0.26 | 0.15 | 0.13 | 0.05–0.27 |
Figure 1.
Bayesian mtDNA gene tree for rough periwinkles. A maximum clade credibility tree was obtained from BEAST analysis. Numbers above and below each node indicate mean node age and posterior probability, respectively. Grey error bars indicate 95% high probability densities for node ages. Posterior probabilities less than 80% are not shown. Geographical regions containing each clade are listed to the right of the figure. Grey boxes surround clades not containing L. saxatilis. For simplicity, this tree was created using unique haplotypes only; however, values included refer to analyses using the full dataset (topologies were identical).
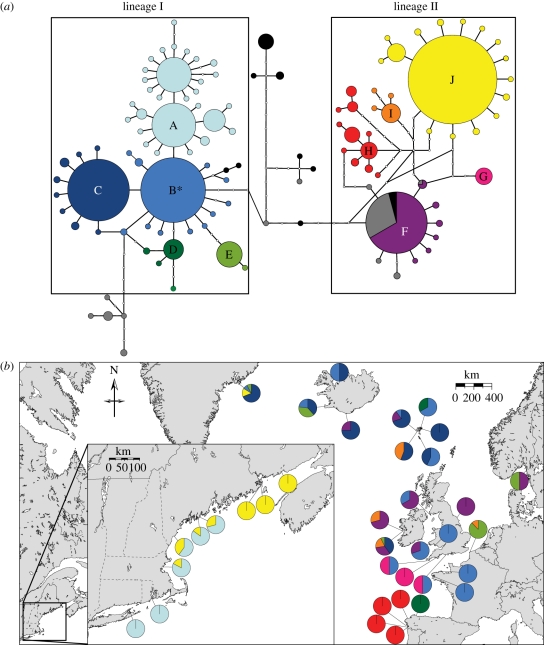
Figure 2.
(a) Maximum parsimony network for rough periwinkles. Lines indicate 95% connection limit and the asterisk denotes the maximum-likelihood root haplotype. Grey indicates L. arcana: black indicates L. compressa. All L. saxatilis mtDNA haplotypes are coloured and letters indicate haplogroups referred to in the text. Circle size is proportional to the number of individuals, with the smallest circles representing one and the largest representing 66. Small white circles denote putative intermediate haplotypes not recovered in our sampling. (b) Haplotype frequencies for L. saxatilis collected at each site (electronic supplementary material, table S1). Colours correspond to (a).
Lineage II, with a TMRCA of 0.45 Ma (0.23–0.73, table 1), is older and more diverse (figure 2a) than lineage I, with a TMRCA of 0.32 Ma (0.15–0.53). Within these lineages, there is further geographical subdivision: western North Atlantic haplotypes (haplogroups A and J) fall into well-supported clades within lineages I and II, respectively, as do Iberian Peninsula haplotypes (haplogroups D and H). Remaining haplotypes are shared broadly across central and northeastern North Atlantic populations.
(c). Phylogeography
The ranges of both L. saxatilis lineages (I and II) overlap broadly across the North Atlantic, but geographical subdivision is evident, and patterns of overlap differ among regions (eastern, central and western) and between basal and derived haplotypes (figure 2). Basal haplotypes of both lineages I (haplogroup B) and II (haplogroup F) are present across the northeastern and central North Atlantic, in Britain, the Faroe Islands and Iceland. Lineage I basal haplotypes (haplogroup B) are also found in France and Greenland, whereas Ireland and Sweden contain basal lineage II haplotypes (haplogroup F).
Derived lineage I and II haplotypes are also present across the central and northeastern North Atlantic. Haplogroup C (lineage I) has a northern distribution, across Ireland, the Faroe Islands, Iceland and Greenland, whereas haplogroup E (lineage I) is found primarily on North Sea shores (France and Sweden), but also in Ireland, Iceland and Greenland. Lineage II-derived haplogroup I is spread among France, Ireland and the Faroe Islands, whereas the divergent haplotype G is localized to southwestern Britain.
All four haplotypes recovered from Greenland are also common across the northeastern (main haplotypes C, B and E) or northwestern (J) North Atlantic (i.e. there are no private haplotypes). The same three northeastern North Atlantic haplotypes (C, B and E) are also found in Iceland, along with two private haplotypes (in haplogroups C and F).
At the southeastern edge of our sampling range, the Iberian Peninsula contains derived haplotypes from both lineages I (haplogroup D) and II (haplogroup H). The diversity within H is much deeper than other haplogroups, with a TMRCA of 0.28 Ma (0.13–0.48). Unlike the rest of the eastern North Atlantic, 13 out of 14 Iberian Peninsula haplotypes are exclusive to this region (i.e. private haplotypes); the 14th Iberian haplotype (D) was also recovered from the Faroe Islands. The ranges of lineages I and II do not overlap on the Iberian Peninsula: lineage I (haplogroup D) is found on Spain's Biscay Coast, while lineage II (haplogroup H) is restricted to the North Atlantic coast.
The western North Atlantic is populated with derived haplotypes from both lineages I and II, and all haplotypes but one (J, also present in Greenland) are unique to this region. The range of lineage I (haplogroup A) extends from south of Cape Cod throughout the southern Gulf of Maine, whereas lineage II (haplogroup J) spans the southern and northern Gulf of Maine. Although their ranges overlap throughout the southern Gulf of Maine (figure 2b, inset), haplogroups A and J are more closely related to central and eastern North Atlantic haplotypes than they are to each other (figure 2a). Western North Atlantic haplotypes from lineage I (haplogroup A), which are found predominantly in southern populations in the western North Atlantic, show greater diversity than those from lineage II (haplogroup J).
(d). Skyride analysis
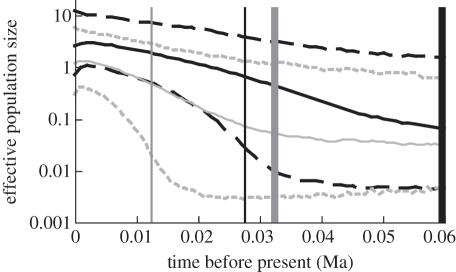
GMRF Skyride analyses show no extreme fluctuations in population size over time, and Bayes factor comparisons reveal that demographic histories do not differ significantly from a constant population size model. We present the data for the western North Atlantic only (figure 3) because inferred past population dynamics for lineages I and II show similar patterns across the North Atlantic. Lineage I (haplogroup A) shows a consistently higher estimate of effective population size over time and TMRCA estimates are slightly older, using both the constant population size model (0.19 Ma, 0.07–0.35) and the more conservative Skyride model (0.06 Ma, 0.03–0.10). Lineage II (haplogroup J) has younger, though overlapping, estimates of TMRCA (0.14 Ma, 0.05–0.26, and 0.03 Ma, 0.01–0.06, respectively).
Figure 3.
GMRF Bayesian Skyride plot of effective population size over time for lineages I and II on western North Atlantic shores. Lineage I (haplogroup A) is shown in black and lineage II (haplogroup J) is shown in grey. Solid and dashed traces represent the median and 95% HPD values for effective population size, respectively. Thick vertical lines indicate mean TMRCA (estimated under a population growth model), while thin vertical lines show the lower 95% HPD TMRCA.
4. Discussion
(a). Gene tree
Our mtDNA gene tree revealed surprising cryptic diversity and a complex evolutionary history for the three North Atlantic rough periwinkles—L. compressa, L. arcana and L. saxatilis. Our results uphold basic relationships within the accepted phylogeny [15], although speciation events appear more recent than previously estimated [43]. The most basal mtDNA lineage, predominantly composed of L. compressa and exclusive to egg-laying species, diverged at approximately 0.92 Ma, coinciding with both the middle Pleistocene transition (1.25–0.70 Ma)—a period of intensification in glacial activity [46]—and the estimated time of divergence between flat periwinkles L. obtusata and L. fabalis [47]. Although L. saxatilis is a brooder [15], identical haplotype and extensive lineage sharing with egg-laying L. arcana and L. compressa were observed and have been previously documented [48–50]. Because L. saxatilis and L. arcana can hybridize (though not with L. compressa [51,52]), haplotype sharing may be due to recent introgressive hybridization. However, shared ancestral variation is more likely [49,50] because (i) the shared haplotype (F) is basal, (ii) no shared haplotypes are observed at sites where the species were sampled together (electronic supplementary material, table S1) and (iii) a more complete complement of possible haplotype diversity is not shared across the sympatric ranges of the three species (unlike L. obtusata and L. fabalis [53]).
Our data indicate that the two mtDNA lineages (I and II), which contain all L. saxatilis (i.e. brooding) individuals sampled, began to diverge from a common ancestor that they shared with the exclusively egg-laying lineage around 0.92 Ma. Because the ranges of both sister species (L. arcana and L. compressa) are limited to the northeastern North Atlantic [15] and the basal diversity in both L. saxatilis lineages I (haplogroup B) and II (haplogroup F) is centred around the British Isles (figure 2), it is most likely that both brooding L. saxatilis lineages arose in the northeastern North Atlantic. Phylogeographic patterns suggest that, following divergence, both L. saxatilis lineages radiated outward, reaching their current amphi-Atlantic distributions. Both nucleotide diversity and TMRCA indicate that range expansion may have occurred approximately 130 Kyr earlier for lineage II (approx. 0.45 Ma) than lineage I (approx. 0.32 Ma).
(b). Phylogeography
Phylogeographic differences in both distributions of basal and derived haplotypes and patterns of lineage overlap across the range of surveyed L. saxatilis populations (Iberian Peninsula, northeastern, central and western North Atlantic) probably result from regional variation in historical glacial processes. The Iberian Peninsula displays unique genetic patterns, including independent ranges of lineages I and II, a high frequency of private haplotypes, relatively high haplotype diversity and population subdivision with isolation by distance [34], which indicate relative population isolation and stability [54]. Contrary to previous hypotheses [34], our data indicate that the Iberian Peninsula was colonized (from the northeastern North Atlantic) at least twice, once by each mtDNA lineage (I and II). Bayesian estimates suggest that most L. saxatilis populations on the Iberian Peninsula have been isolated from remaining L. saxatilis since lineage II colonized the Atlantic coast of the Iberian Peninsula at approximately 0.28 Ma. For other taxa, the Bay of Biscay has also served as an isolating barrier (e.g. L. obtusata and L. fabalis [53], Ascophyllum nodosum [14]) and prevented the recolonization of the northeastern North Atlantic by Iberian Peninsula genetic stocks during interglacial periods (e.g. Fucus serratus [9]).
In the northeastern North Atlantic, from the Brittany Peninsula to the Faroe Islands, the phylogeographic pattern is the most complex and thus difficult to interpret. It has been hypothesized that the geographical complexity of this region's shoreline, and repeated glacial disturbance owing to ice cover and sea-level change, resulted in many glacial refugia, including the English Channel/Hurd Deep, southwestern Ireland, the Faroe Islands and northern Norway [3]. The broad distributions of lineage I and II basal haplogroups (B and F, respectively) suggest multiple glacial refugia throughout the Pleistocene. Lineage I haplogroup C, which is distributed across the Faroe Islands, Iceland, Greenland and southwestern Ireland, probably diverged in a more northern refugium, perhaps the Faroe Islands, but may also have survived in southwestern Ireland. Lineage II haplogroup I shows a similar pattern, whereas lineage II haplogroup E may have recolonized the North Sea from a different northern refugium (e.g. Norway [3]). Localization of haplotype G to southwestern Britain supports a marine refugium in the Hurd Deep during the LGM [7,14], while the extent of native diversity in the Faroe Islands (nine private haplotypes) provides evidence that they did indeed serve as a marine refugium during the LGM [54–57] (but see Ingolfsson [10,58]).
Our data suggest that the two central North Atlantic (Iceland and Greenland) populations of L. saxatilis have experienced the highest levels of glacial disturbance, probably including local extinction. Similar to many marine species [2,10], low haplotype diversity in this region indicates that L. saxatilis recolonized it recently, probably following the LGM, and phylogeographic patterns show that recolonization proceeded predominantly from the northeastern North Atlantic. A subset of the variation found in the Faroe Islands, Ireland and the North Sea (haplotypes C, B and E) was found in both Iceland and Greenland. The presence of two private haplotypes in Iceland, while Greenland lacked private haplotypes, indicates that recolonization probably occurred via east-to-west stepping-stone migration. Interestingly, Greenland also appears to have been recolonized from northwestern North Atlantic populations (haplotype J).
For the western North Atlantic, our data suggest three key findings: (i) this region was colonized by L. saxatilis in two separate trans-Atlantic migration events; (ii) contrary to previous hypotheses [15,59], western North Atlantic L. saxatilis survived the LGM; and (iii) lineages I and II survived in separate southern and northern glacial refugia, respectively. Lack of trans-Atlantic haplotype sharing in either lineage demonstrates that neither lineage I nor II colonized the western North Atlantic following the LGM [2]. TMRCA and haplotype diversity indicate that lineage I (haplogroup A) arrived on western North Atlantic shores prior to lineage II (haplogroup J), possibly as long ago as 0.32 Ma, but probably around 0.19 Ma, during the second to last interglacial period. TMRCA indicates that lineage II (haplogroup J) may have arrived during the last interglacial period. However, as haplogroup J is more distantly related to the rest of lineage II than haplogroup A is to lineage I, haplogroup J may have been isolated from eastern North Atlantic lineage II earlier than we estimated. Haplogroup A appears to have survived the LGM south of the Laurentide ice sheet [2,11], and Skyride analysis indicates that effective population size remained relatively high throughout the LGM (figure 3). Haplogroup J probably survived in a northern location, providing additional evidence for a periglacial refugium in North Atlantic Canada [3,7,11–14]. Since the termination of the LGM (approx. 18 Ka), both populations appear to have expanded and now share a zone of secondary contact in the southern Gulf of Maine [3]. It is likely that the brooding life-history strategy [60] and flexible habitat requirements [15] allowed L. saxatilis to survive drastic climatic change and rocky habitat limitation associated with the LGM, whereas many rocky intertidal organisms suffered local extinction (e.g. L. obtusata and N. lapillus [2,4,8]). Lack of extreme population fluctuations in either lineage over this time period (figure 3) indicates that the LGM did not severely impact genetic diversity but instead drove the distribution of genetic variation (e.g. [14]) in western North Atlantic L. saxatilis.
(c). Conclusions
The existence of two distinct mtDNA lineages, which now show strong phylogeographic structure across the trans-Atlantic range of L. saxatilis, demonstrates that this species has had a complex evolutionary history shaped by repeated glacial cycles and persistence in multiple glacial refugia. This broad phylogeographic context also provides a framework for examination of patterns of ecological differentiation and incipient speciation in L. saxatilis. Future work must examine the relationship between mtDNA lineage and nuclear gene exchange (i.e. AFLP, SNP or microsatellites) across populations and among L. saxatilis ecotypes. Britain, Sweden and the region between Spanish Atlantic and Biscay coasts will be of particular interest in defining the relative roles of and/or interaction between sympatric and allopatric processes, because our data indicate that genetic differentiation may be ongoing at more than one scale in the L. saxatilis system. In addition, a further focus on western North Atlantic L. saxatilis is needed. Significant spatial and temporal separation of lineages I and II, with recent (less than 20 Ka) secondary contact in the southern Gulf of Maine, make this the ideal region for detailed examination of interactions between lineages. Identification of reduced nuclear gene flow or reproductive isolation between mtDNA lineages could lead to redefinition of the species L. saxatilis.
Acknowledgements
We thank C.S. Wilding for use of his collections, E. Bryson, R. Dolecal and C. Matassa for collection assistance, D. Reid, S. Williams, R. Butlin and J. Galindo for helpful conversations, and E. Bryson, S. Donelan, L. Hemond, S. Kent, L. Miller, C. Matassa, K. McClure, C. Sorte and four anonymous reviewers for comments on earlier versions of the manuscript. This work was supported under a National Science Foundation Graduate Research Fellowship and a Marie Curie Fellowship for Early Stage Research Training to M.M.D., and NSF grants to G.C.T. (OCE-0648525, OCE-0727628) and S.V.V. (OCE-0848345). This is Contribution No. 279 from the Marine Science Center.
References
- 1.Vermeij G. J. 1991. Anatomy of an invasion: the trans-Arctic interchange. Paleobiology 17, 281–307 [Google Scholar]
- 2.Wares J. P., Cunningham C. W. 2001. Phylogeography and historical ecology of the north Atlantic intertidal. Evolution 55, 2455–2469 [DOI] [PubMed] [Google Scholar]
- 3.Maggs C. A., et al. 2008. Evaluating signatures of glacial refugia for north Atlantic benthic marine taxa. Ecology 89, S108–S122 10.1890/08-0257.1 (doi:10.1890/08-0257.1) [DOI] [PubMed] [Google Scholar]
- 4.Cunningham C. W., Collins T. M. 1998. Beyond area relationships: extinction and recolonization in molecular marine biogeography. In Molecular ecology and evolution: approaches and applications (eds Schierwater, Streit B., Wagner G., DeSalle R.), pp. 297–321 Basel, Switzerland: Birkhauser Verlag [Google Scholar]
- 5.Gladenkov A. Y., Oleinik A. E., Marincovich L., Barinov K. B. 2002. A refined age for the earliest opening of Bering Strait. Palaeogeogr. Palaeoclimat. Palaeoecol. 183, 321–328 10.1016/S0031-0182(02)00249-3 (doi:10.1016/S0031-0182(02)00249-3) [DOI] [Google Scholar]
- 6.Coyer J. A., Peters A. F., Stam W. T., Olsen J. L. 2003. Post-ice age recolonization and differentiation of Fucus serratus L. (Phaeophyceae; Fucaceae) populations in Northern Europe. Mol. Ecol. 12, 1817–1829 10.1046/j.1365-294X.2003.01850.x (doi:10.1046/j.1365-294X.2003.01850.x) [DOI] [PubMed] [Google Scholar]
- 7.Provan J., Wattier R. A., Maggs C. A. 2005. Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Mol. Ecol. 14, 793–803 10.1111/j.1365-294X.2005.02447.x (doi:10.1111/j.1365-294X.2005.02447.x) [DOI] [PubMed] [Google Scholar]
- 8.Colson I., Hughes R. N. 2007. Contrasted patterns of genetic variation in the dogwhelk Nucella lapillus along two putative post-glacial expansion routes. Mar. Ecol. Prog. Ser. 343, 183–191 10.3354/meps06879 (doi:10.3354/meps06879) [DOI] [Google Scholar]
- 9.Hoarau G., Coyer J. A., Veldsink J. H., Stam W. T., Olsen J. L. 2007. Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Mol. Ecol. 16, 3606–3616 10.1111/j.1365-294X.2007.03408.x (doi:10.1111/j.1365-294X.2007.03408.x) [DOI] [PubMed] [Google Scholar]
- 10.Ingolfsson A. 1992. The origin of the rocky shore fauna of Iceland and the Canadian Maritimes. J. Biogeogr. 19, 705–712 10.2307/2845711 (doi:10.2307/2845711) [DOI] [Google Scholar]
- 11.Wares J. P. 2002. Community genetics in the Northwestern Atlantic intertidal. Mol. Ecol. 11, 1131–1144 10.1046/j.1365-294X.2002.01510.x (doi:10.1046/j.1365-294X.2002.01510.x) [DOI] [PubMed] [Google Scholar]
- 12.Chopin T., Bird C. J., Murphy C. A., Osborne J. A., Patwary M. U., Floc'h J.-Y. 1996. A molecular investigation of polymorphism in the North Atlantic red alga Chondrus crispus (Gigartinales). Phycol. Res. 44, 69–80 10.1111/j.1440-1835.1996.tb00378.x (doi:10.1111/j.1440-1835.1996.tb00378.x) [DOI] [Google Scholar]
- 13.Bigg G. R., Cunningham C. W., Ottersen G., Pogson G. H., Wadley M. R., Williamson P. 2008. Ice-age survival of Atlantic cod: agreement between palaeoecology models and genetics. Proc. R. Soc. B 275, 163–173 10.1098/rspb.2007.1153 (doi:10.1098/rspb.2007.1153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olsen J. L., Zechman F. W., Hoarau G., Coyer J. A., Stam W. T., Valero M., Åberg P. 2010. The phylogeographic architecture of the fucoid seaweed Ascophyllum nodosum: an intertidal ‘marine tree’ and survivor of more than one glacial-interglacial cycle. J. Biogeogr. 37, 842–856 10.1111/j.1365-2699.2009.02262.x (doi:10.1111/j.1365-2699.2009.02262.x) [DOI] [Google Scholar]
- 15.Reid D. G. 1996. Systematics and evolution of Littorina. London, UK: Ray Society [Google Scholar]
- 16.Reid D. G. 1990. Trans-Arctic migration and speciation induced by climatic change: the biogeography of Littorina (Mollusca: Gastropoda). Bull. Mar. Sci. 47, 35–49 [Google Scholar]
- 17.Johannesson K. 2001. Parallel speciation: a key to sympatric divergence. Trends Ecol. Evol. 16, 148–153 10.1016/S0169-5347(00)02078-4 (doi:10.1016/S0169-5347(00)02078-4) [DOI] [PubMed] [Google Scholar]
- 18.Johannesson K. 2003. Evolution in Littorina: ecology matters. J. Sea Res. 49, 107–117 10.1016/S1385-1101(02)00218-6 (doi:10.1016/S1385-1101(02)00218-6) [DOI] [Google Scholar]
- 19.Janson K. 1983. Selection and migration in two distinct phenotypes of Littorina saxatilis in Sweden. Oecologia 59, 58–61 10.1007/BF00388072 (doi:10.1007/BF00388072) [DOI] [PubMed] [Google Scholar]
- 20.Johannesson K., Rolan-Alvarez E., Ekendahl A. 1995. Incipient reproductive isolation between two sympatric morphs of the intertidal snail Littorina saxatilis. Evolution 49, 1180–1190 10.2307/2410443 (doi:10.2307/2410443) [DOI] [PubMed] [Google Scholar]
- 21.Hull S. L., Grahame J. W., Mill P. J. 1996. Morphological divergence and evidence for reproductive isolation in Littorina saxatilis (Olivi) in northeast England. J. Mollus. Stud. 62, 89–99 10.1093/mollus/62.1.89 (doi:10.1093/mollus/62.1.89) [DOI] [Google Scholar]
- 22.Wilding C. S., Butlin R. K., Grahame J. W. 2001. Differential gene exchange between parapatric morphs of Littorina saxatilis detected using AFLP markers. J. Evol. Biol. 14, 611–619 10.1046/j.1420-9101.2001.00304.x (doi:10.1046/j.1420-9101.2001.00304.x) [DOI] [Google Scholar]
- 23.Grahame J. W., Wilding C. S., Butlin R. K. 2006. Adaptation to a steep environmental gradient and an associated barrier to gene exchange in Littorina saxatilis. Evolution 60, 268–278 [PubMed] [Google Scholar]
- 24.Panova M., Hollander J., Johannesson K. 2006. Site-specific genetic divergence in parallel hybrid zones suggests nonallopatric evolution of reproductive barriers. Mol. Ecol. 15, 4021–4031 10.1111/j.1365-294X.2006.03067.x (doi:10.1111/j.1365-294X.2006.03067.x) [DOI] [PubMed] [Google Scholar]
- 25.Galindo J., Moran P., Rolan-Alvarez E. 2009. Comparing geographical genetic differentiation between candidate and noncandidate loci for adaptation strengthens support for parallel ecological divergence in the marine snail Littorina saxatilis. Mol. Ecol. 18, 919–930 10.1111/j.1365-294X.2008.04076.x (doi:10.1111/j.1365-294X.2008.04076.x) [DOI] [PubMed] [Google Scholar]
- 26.Rolan-Alvarez E. 2007. Sympatric speciation as a by-product of ecological adaptation in the Galician Littorina saxatilis hybrid zone. J. Mollus. Stud. 73, 1–10 10.1093/mollus/eyl023 (doi:10.1093/mollus/eyl023) [DOI] [Google Scholar]
- 27.Hollander J., Lindegarth M., Johannesson K. 2005. Local adaptation but not geographical separation promotes assortative mating in a snail. Anim. Behav. 70, 1209–1219 10.1016/j.anbehav.2005.03.014 (doi:10.1016/j.anbehav.2005.03.014) [DOI] [Google Scholar]
- 28.Butlin R. K., Galindo J., Grahame J. W. 2008. Sympatric, parapatric or allopatric: the most important way to classify speciation? Phil. Trans. R. Soc. B 363, 2997–3007 10.1098/rstb.2008.0076 (doi:10.1098/rstb.2008.0076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munday P. L., van Herwerden L., Dudgeon C. L. 2004. Evidence for sympatric speciation by host shift in the sea. Curr. Biol. 14, 1498–1504 10.1016/j.cub.2004.08.029 (doi:10.1016/j.cub.2004.08.029) [DOI] [PubMed] [Google Scholar]
- 30.Feder J. L., et al. 2003. Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl Acad. Sci. USA 100, 10 314–10 319 10.1073/pnas.1730757100 (doi:10.1073/pnas.1730757100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolnick D. I., Fitzpatrick B. M. 2007. Sympatric speciation: models and empirical evidence. Annu. Rev. Ecol., Evol. Syst. 38, 459–487 10.1146/annurev.ecolsys.38.091206.095804 (doi:10.1146/annurev.ecolsys.38.091206.095804) [DOI] [Google Scholar]
- 32.Winnepenninckx B., Backeljau T., De Wachter R. 1993. Extraction of high molecular weight DNA from molluscs. Trends Genet. 9, 407. 10.1016/0168-9525(93)90102-N (doi:10.1016/0168-9525(93)90102-N) [DOI] [PubMed] [Google Scholar]
- 33.Wilding C. S., Mill P. J., Grahame J. W. 1999. Partial sequence of the mitochondrial genome of Littorina saxatilis: relevance to gastropod phylogenetics. J. Mol. Evol. 48, 348–359 10.1007/PL00006479 (doi:10.1007/PL00006479) [DOI] [PubMed] [Google Scholar]
- 34.Quesada H., Posada D., Caballero A., Moran P., Rolan-Alvarez E. 2007. Phylogenetic evidence for multiple sympatric ecological diversification in a marine snail. Evolution 61, 1600–1612 10.1111/j.1558-5646.2007.00135.x (doi:10.1111/j.1558-5646.2007.00135.x) [DOI] [PubMed] [Google Scholar]
- 35.Librado P., Rozas J. 2009. DnaSP v. 5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 10.1093/bioinformatics/btp187 (doi:10.1093/bioinformatics/btp187) [DOI] [PubMed] [Google Scholar]
- 36.Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Y. X., Li W. H. 1993. Statistical tests of neutrality of mutations. Genetics 133, 693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clement M., Posada D., Crandall K. A. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659 10.1046/j.1365-294x.2000.01020.x (doi:10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- 39.Drummond A., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nylander J. A. A. MrModeltest v. 2. 2004. See http://www.abc.se/∼nylander . [Google Scholar]
- 41.Drummond A. J., Ho S. Y. W., Phillips M. J., Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88. 10.1371/journal.pbio.0040088 (doi:10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suchard M. A., Weiss R. E., Sinsheimer J. S. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18, 1001–1013 [DOI] [PubMed] [Google Scholar]
- 43.Reid D. G., Rumbak E., Thomas R. H. 1996. DNA, morphology, and fossils: phylogeny and evolutionary rates of the gastropod genus Littorina. Phil. Trans. R. Soc. Lond. B 351, 877–895 10.1098/rstb.1996.0082 (doi:10.1098/rstb.1996.0082) [DOI] [PubMed] [Google Scholar]
- 44.Minin V. N., Bloomquist E. W., Suchard M. A. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 25, 1459–1471 10.1093/molbev/msn090 (doi:10.1093/molbev/msn090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rambaut A., Drummond A. J. 2009. Tracer v. 1.5. See http://beast.bio.ed.ac.uk/Tracer [Google Scholar]
- 46.Clark P. U., Archer D., Pollard D., Blum J. D., Rial J. A., Brovkin V., Mix A. C., Pisias N. G., Roy M. 2006. The middle Pleistocene transition: characteristics, mechanisms, and implications for long-term changes in atmospheric pCO2. Q. Sci. Rev. 25, 3150–3184 10.1016/j.quascirev.2006.07.008 (doi:10.1016/j.quascirev.2006.07.008) [DOI] [Google Scholar]
- 47.Tatarenkov A. 1995. Genetic-divergence between sibling species Littorina mariae Sacchi and Rastelli and Littorina obtusata (L.) (Mollusca, Gastropoda) from the White Sea. Ophelia 40, 207–218 [Google Scholar]
- 48.Small M. P., Gosling E. M. 2000. Genetic structure and relationships in the snail species complex Littorina arcana Hannaford Ellis, L. compressa Jeffreys and L. saxatilis (Olivi) in the British Isles using SSCPs of cytochrome-b fragments. Heredity 84, 692–701 10.1046/j.1365-2540.2000.00717.x (doi:10.1046/j.1365-2540.2000.00717.x) [DOI] [PubMed] [Google Scholar]
- 49.Wilding C. S., Grahame J., Mill P. J. 2000. Mitochondrial DNA Col haplotype variation in sibling species of rough periwinkles. Heredity 85, 62–74 10.1046/j.1365-2540.2000.00718.x (doi:10.1046/j.1365-2540.2000.00718.x) [DOI] [PubMed] [Google Scholar]
- 50.Wilding C. S., Grahame J., Mill P. J. 2000. Nuclear DNA restriction site polymorphisms and the phylogeny and population structure of an intertidal snail species complex (Littorina). Hereditas 133, 9–18 10.1111/j.1601-5223.2000.00009.x (doi:10.1111/j.1601-5223.2000.00009.x) [DOI] [PubMed] [Google Scholar]
- 51.Warwick T., Knight A. J., Ward R. D. 1990. Hybridization in the Littorina saxatilis species complex (Prosobranchia: Mollusca). Hydrobiologia 193, 109–116 10.1007/BF00028070 (doi:10.1007/BF00028070) [DOI] [Google Scholar]
- 52.Mikhailova N., Gracheva Y., Backeljau T., Granovitch A. 2009. A potential species-specific molecular marker suggests interspecific hybridization between sibling species Littorina arcana and L. saxatilis (Mollusca, Caenogastropoda) in natural populations. Genetica 137, 333–340 10.1007/s10709-009-9397-4 (doi:10.1007/s10709-009-9397-4) [DOI] [PubMed] [Google Scholar]
- 53.Kemppainen P., Panova M., Hollander J., Johannesson K. 2009. Complete lack of mitochondrial divergence between two species of NE Atlantic marine intertidal gastropods. J. Evol. Biol. 22, 2000–2011 10.1111/j.1420-9101.2009.01810.x (doi:10.1111/j.1420-9101.2009.01810.x) [DOI] [PubMed] [Google Scholar]
- 54.Lessa E. P., Cook J. A., Patton J. L. 2003. Genetic footprints of demographic expansion in North America, but not Amazonia, during the Late Quaternary. Proc. Natl Acad. Sci. USA 100, 10331–10334 10.1073/pnas.1730921100 (doi:10.1073/pnas.1730921100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahlgren T. G., Weinberg J. R., Halanych K. M. 2000. Phylogeography of the ocean quahog (Arctica islandica): influences of paleoclimate on genetic diversity and species range. Am. Zool. 40, 989–989 [Google Scholar]
- 56.Wares J. P. 2001. Intraspecific variation and geographic isolation in Idotea balthica (Isopoda: Valvifera). J. Crust. Biol. 21, 1007–1013 10.1651/0278-0372(2001)021[1007:IVAGII]2.0.CO;2 (doi:10.1651/0278-0372(2001)021[1007:IVAGII]2.0.CO;2) [DOI] [Google Scholar]
- 57.Roman J., Palumbi S. R. 2004. A global invader at home: population structure of the green crab, Carcinus maenas, in Europe. Mol. Ecol. 13, 2891–2898 10.1111/j.1365-294X.2004.02255.x (doi:10.1111/j.1365-294X.2004.02255.x) [DOI] [PubMed] [Google Scholar]
- 58.Ingolfsson A. 2009. A marine refugium in Iceland during the last glacial maximum: fact or fiction? Zool. Scr. 38, 663–665 10.1111/j.1463-6409.2009.00405.x (doi:10.1111/j.1463-6409.2009.00405.x) [DOI] [Google Scholar]
- 59.Blakeslee A. M. H., Byers J. E. 2008. Using parasites to inform ecological history: comparisons among three congeneric marine snails. Ecology 89, 1068–1078 10.1890/07-0832.1 (doi:10.1890/07-0832.1) [DOI] [PubMed] [Google Scholar]
- 60.Johannesson K. 1988. The paradox of Rockall: why is a brooding gastropod (Littorina saxatilis) more widespread than one having a planktonic larval dispersal stage (L. littorea)? Mar. Biol. 99, 507–513 10.1007/BF00392558 (doi:10.1007/BF00392558) [DOI] [Google Scholar]