Figure 2.
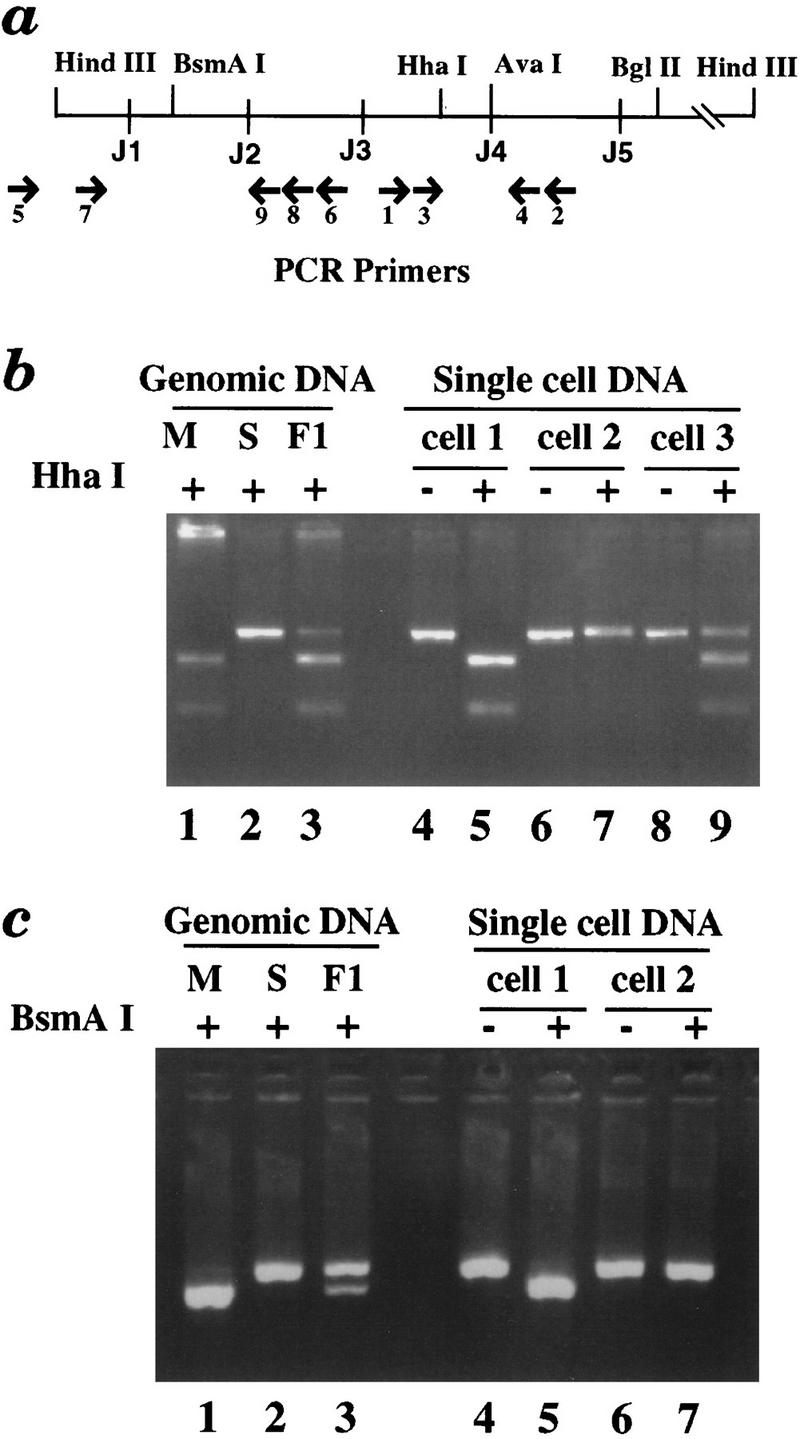
MSRE–PCR analysis of the κ gene in single cells. (a) A map of the Jκ region showing the location of primers used for MSRE–PCR analysis and relevant restriction enzyme sites. AvaI was used to detect DNA methylation, whereas the polymorphic HhaI and BsmAI were employed to distinguish between the two κ alleles. (b) (Lanes 1–3) As a control, genomic DNAs from M. musculus, M. spretus, or F1 mice were PCR amplified with primers P3 and P4, digested with HhaI (used here because of a polymorphism, not because of its methyl-sensitivity), and analyzed on a 4% agarose gel. The M. musculus gene is digested, the M. spretus gene does not cut, and the F1 DNA yields a product that shows 50% digestion. (Lanes 4–7) DNA from two single κ+ B cells was digested with AvaI and PCR amplified. (The secondary PCR primers were P1 and P2.) Uncut PCR products are in lanes 4 and 6; HhaI digested PCR products are in lanes 5 and 7. Cells 1 and 2 are monoallelic (M. musculus and M. spretus allele, respectively). Most κ+ B cells analyzed were monoallelic. (Lanes 8, 9) The same analysis of a single midbrain cell (cell 3). Both alleles were amplified from this cell and in most of the control cells analyzed. (c) (Lanes 1–3) Control M. musculus, M. spretus, or F1 genomic DNAs were PCR amplified by primers P7 and P9, digested with BsmAI, and analyzed on a 4% agarose gel. (Lanes 4–7) Tertiary PCR products derived from cells 1 and 2 (from b) using primers P7 and P9. The tertiary amplification signal is dependent on the germ-line-specific primary PCR (using primers P5 and P6). Uncut PCR products are in lanes 4 and 6, and corresponding BsmAI-digested PCR products are in lanes 5 and 7. Cell 1 (shown previously to have a methylated M. musculus allele) has an unrearranged M. musculus allele; cell 2 (shown previously to have a methylated M. spretus allele) has an unrearranged M. spretus allele. All together, 15 individual cells with PCR products from both regions were analyzed.
