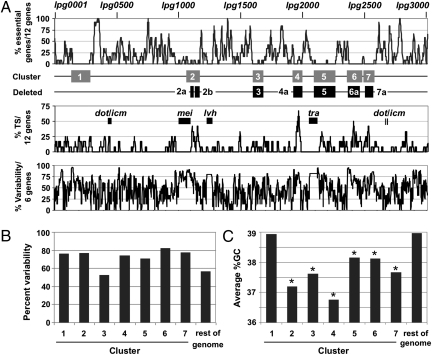
Fig. 2.
Mosaic architecture of the L. pneumophila genome. (A) Seven genomic islands are devoid of genes essential for growth in rich media but exhibit a high concentration of Dot/Icm TS. (Top) Running average plot of the percentage of essential genes per 12 genes. The seven gene clusters identified (gray boxes) and the six genomic loci deleted (black boxes) are indicated. (Middle) Running average plot of the percentage of Dot/Icm TS per 12 genes. The locations of the dot/icm, tra, lvh, and mei loci (black boxes) are individually labeled. (Bottom) Running average plot of the percent variability per 6 genes. Pairwise comparison of the L. pneumophila Philadelphia 1 genome to Paris, Lens, Corby, and Alcoy strains was used to identify genes that are conserved or absent across all five species. Percent variability is the number of genes inserted or deleted relative to the total number of genes in that region. (B) Six of the genomic islands have a higher percent variability than the rest of the genome, as defined in A, compared with other sequenced strains of L. pneumophila. (C) Six genomic islands exhibit a lower average %GC for all genes in the cluster compared with the rest of the genome. Clusters exhibiting a statistically significant difference based on a two-tailed Student t test P value <0.05 relative to the rest of the genome are indicated by an asterisk. %GC, percent of guanine and cytosine bases.