Abstract
Hemagglutinin (HA) and neuraminidase (NA) are two surface proteins of influenza viruses which are known to play important roles in the viral life cycle and the induction of protective immune responses1,2. As the main target for neutralizing antibodies, HA is currently used as the influenza vaccine potency marker and is measured by single radial immunodiffusion (SRID)3. However, the dependence of SRID on the availability of the corresponding subtype-specific antisera causes a minimum of 2-3 months delay for the release of every new vaccine. Moreover, despite evidence that NA also induces protective immunity4, the amount of NA in influenza vaccines is not yet standardized due to a lack of appropriate reagents or analytical method5. Thus, simple alternative methods capable of quantifying HA and NA antigens are desirable for rapid release and better quality control of influenza vaccines.
Universally conserved regions in all available influenza A HA and NA sequences were identified by bioinformatics analyses6-7. One sequence (designated as Uni-1) was identified in the only universally conserved epitope of HA, the fusion peptide6, while two conserved sequences were identified in neuraminidases, one close to the enzymatic active site (designated as HCA-2) and the other close to the N-terminus (designated as HCA-3)7. Peptides with these amino acid sequences were synthesized and used to immunize rabbits for the production of antibodies. The antibody against the Uni-1 epitope of HA was able to bind to 13 subtypes of influenza A HA (H1-H13) while the antibodies against the HCA-2 and HCA-3 regions of NA were capable of binding all 9 NA subtypes. All antibodies showed remarkable specificity against the viral sequences as evidenced by the observation that no cross-reactivity to allantoic proteins was detected. These universal antibodies were then used to develop slot blot assays to quantify HA and NA in influenza A vaccines without the need for specific antisera7,8. Vaccine samples were applied onto a PVDF membrane using a slot blot apparatus along with reference standards diluted to various concentrations. For the detection of HA, samples and standard were first diluted in Tris-buffered saline (TBS) containing 4M urea while for the measurement of NA they were diluted in TBS containing 0.01% Zwittergent as these conditions significantly improved the detection sensitivity. Following the detection of the HA and NA antigens by immunoblotting with their respective universal antibodies, signal intensities were quantified by densitometry. Amounts of HA and NA in the vaccines were then calculated using a standard curve established with the signal intensities of the various concentrations of the references used.
Given that these antibodies bind to universal epitopes in HA or NA, interested investigators could use them as research tools in immunoassays other than the slot blot only.
Protocol
1. Preparation of reagents and equipment
Before starting the slot blot procedure, prepare 20_mls of 4M urea solution in Tris-buffered saline (20 mM Tris, 137 mM NaCl, pH 7.6) (TBS) for the hemagglutinin, or HA, slot blot, or 20_mls of a 0.01% Zwittergent solution in TBS for the neuraminidase, or NA, slot blot. While 4M urea should be prepared fresh every time, a 10% Zwittergent stock solution in dH2O is stable for at least 6 months at room temperature and can therefore be diluted to 0.01% in TBS before each assay.
Prepare 2000 mls of washing buffer by adding Tween-20 to a TBS solution to a final concentration of 0.1%. Prepare 80 mls of blocking buffer by dissolving skim milk powder (blotting grade non-fat dry milk) to a final concentration of 5% w/v in washing buffer.
Activate the PVDF membrane (pre-cut to a 9 x 12 cm size) in methanol for 15 seconds, followed by a 2 minutes rinse in ddH2O. Soak the membrane in TBS until use. Pre-wet 3 sheets of Bio-Dot SF filter paper in TBS until used.
2. Preparation of influenza vaccine samples and reference standard for HA slot blot
Influenza vaccine reference standards with pre-determined HA content corresponding to the vaccine strains to be tested were obtained from the Centre for Biologics Evaluation and Research (CBER/FDA, USA) or the National Institute for Biological Standard and Control (NIBSC, UK) and are used for the HA quantification by slot blot. The investigators could also prepare their own antigen standards using established procedures as described in references 9 and 10.
For HA reference antigens and test vaccine samples: Dilute the HA reference antigen stock in 4M urea/TBS to the following concentrations: 0, 0.0938, 0.1875, 0.375, 0.75, 1.5, 3 and 6 μg HA/ml. Mix well. Then, dilute test vaccine samples for human use obtained from vaccine producers or made in-house for scientific research purposes in 4M urea/TBS and mix well. While duplicates of the reference antigen and vaccine samples can be run, triplicates are preferred. Prepare 3 dilutions of each sample (2-fold difference) to allow the sample concentration to fall within the standard curve. Prepare 450 μl or 650 μl of each reference antigen or test vaccine sample dilution to run duplicates or triplicates respectively. Monovalent influenza A vaccines can be tested with this slot blot method. The HA content of the reference antigens was previously determined using SRID which is the currently used standard test for influenza vaccine preparations. The HA content determined by SRID was used as the initial reference antigen stock HA concentration for the preparation of the dilutions for the slot blot standard curve.
3. Preparation of influenza vaccine samples and reference standard for NA slot blot
Influenza vaccine reference standards corresponding to the vaccine strains to be tested were obtained from the Centre for Biologics Evaluation and Research (CBER/FDA, USA) or the National Institute for Biological Standard and Control (NIBSC, UK) and are used for the NA quantification by slot blot. The NA content of these reference samples was determined by SDS-PAGE analysis of deglycosylated samples in conjunction with densitometry scanning analysis, as described previously9, with slight modifications10. In brief, the deglycosylated vaccine reference samples (5 μg) were mixed with sample buffer and loaded on the gel. The gel was run at 20 mA for approximately 90 min until the tracking dye just run out of the gel, followed by Sypro staining. Densitometry quantification of proteins was carried out using a Fluorchem gel documentation system (Alpha Innotech). The amount of NA was determined based on the ratio of the NA proteins to the total proteins as determined by Lowry assay. Reference samples prepared using this method can then be used in slot blot for the quantification of NA contents in vaccine samples from vaccines manufacturers.
For NA reference antigens and test vaccine samples: Dilute the NA reference antigen stock in 0.01% Zwittergent/TBS to the following concentrations: 0, 0.039, 0.078, 0.1562, 0.3125, 0.625, 1.25, 2.5, 5 and 10 μg NA/ml. Mix well. Then, dilute test vaccine samples for human use obtained from vaccine producers or made in-house for scientific research purposes in 0.01% Zwittergent/TBS and mix well. While duplicates of the reference antigen and vaccine samples can be run, triplicates are preferred. Prepare 3 dilutions of each sample (2-fold difference) to allow the sample concentration to fall within the standard curve. Prepare 450 μl or 650 μl of each reference antigen or test vaccine sample dilution to run duplicates or triplicates respectively. Monovalent influenza A vaccines can be tested with this slot blot method.
4. Assembly of the Bio-Dot SF Microfiltration apparatus and blotting of samples onto PVDF membrane
Assemble a previously cleaned and dried Bio-Dot SF Microfiltration apparatus. Place the gasket support plate into the vacuum manifold and place the sealing gasket on top.Place the 3 moistened sheets of Bio-Dot SF filter paper over the gasket, followed by the pre-soaked PVDF membrane. Place the sample template on top of the membrane and finger-tighten the four screws using a diagonal crossing pattern to ensure uniform application of pressure on the membrane surface.
Verify that the 3-way valve is set to expose the vacuum manifold to the vacuum source only before turning on the pump and connecting the vacuum.
Turn on the pump and connect to the Bio-Dot apparatus. Make sure that the flow valve is positioned at a level below the sample wells for rapid and proper drainage of the samples in the following steps.
Repeat the screw tightening step using the diagonal crossing pattern. Tightening the screws while the vacuum is applied ensures a tighter seal and prevents cross contamination between wells.
Change the flow valve to expose the vacuum manifold to air and turn off the pump.
Apply 100 μl of TBS to all wells using a multichannel pipettor to rehydrate the membrane thus ensuring uniform binding of the antigen. Care must be taken to avoid the formation of bubbles and to apply the solution in the middle of the well, as close to the bottom as possible, in order to cover the slot evenly. Burst any air bubble accidently introduced with a pipettor tip.
Change the flow valve to expose the manifold to both air and the vacuum, turn on the pump, and gently drain the buffer from the wells by putting a finger over the port exposed to air to regulate the amount of vacuum.
Change the flow valve to expose the manifold to air as soon as the buffer has completely drained from all wells. Turn off the pump or disconnect the vacuum.
Apply 200 μl of each standard or vaccine sample dilution per well. Pipet samples one at a time, in the center of each well, as close to the bottom as possible, taking care to avoid introducing air bubbles. Burst any air bubble accidently introduced with a pipettor tip. Put 200 μl of sample dilution buffer (4M urea/TBS or 0.01% Zwittergent/TBS) in unused wells to ensure proper vacuum to the wells in use.
Adjust the flow valve to expose the manifold to both air and the vacuum. Turn on the pump (or reconnect the vacuum) and gently allow the samples to completely drain from the wells by controlling the amount of vacuum with one finger over the port exposed to air. Remove the finger from the port to release the vacuum as soon as all the samples have drained.
Immediately add 200 μl of TBS to each well using a multichannel pipettor. Apply a gentle vacuum to drain and wash the wells by controlling the amount of vacuum with one finger over the port exposed to air.
Repeat the wash step described in step 4.11. As soon as the wells have completely drained, loosen the screws of the sample template and turn off the vacuum to avoid overdrying the wells. Immediately proceed to remove the membrane.
Place the membrane into blocking buffer and incubate overnight at 4°C, with gentle rocking. Use enough blocking buffer to completely cover the membrane.
5. Immunoblotting of HA and NA antigens
Following the overnight incubation in blocking buffer, wash the membrane twice for 5 minutes with TBS/0.1% Tween-20 washing buffer. For all washings and incubations, ensure the membrane is completely covered with solution.
Incubate the membrane with a universal rabbit antibody against HA (such as Uni-1) or NA (such as HCA-2 or HCA-3) diluted in 5% w/v BSA in washing buffer for one hour at room temperature, with gentle rocking. Optimal antibody concentrations need to be determined for each antibody stock. Dilutions of 1:4000 of rabbit anti-serum gave optimal results for the above mentioned antibodies.
Wash the membrane three times, at room temperature with gentle rocking for 15 minutes each time, to remove any unbound primary antibodies.
Incubate the membrane with an HRP-conjugated anti-rabbit secondary antibody. Use a 1:50000 dilution of the Thermo ImmunoPure Goat anti-rabbit IgG, peroxidase conjugated antibody in blocking buffer and incubate at room temperature for 30 minutes with gentle rocking.
Wash the membrane three times, rocking for 15 minutes each time, at room temperature with washing buffer to remove any unbound secondary antibodies.
Wash the membrane with TBS for 5 minutes with gentle rocking to remove residual Tween-20 detergent from the membrane surface.
Prepare the substrate with 3 ml of each reagent from the Supersignal West Dura Extended Duration Substrate kit from Thermo Scientific and mix well.
Place the membrane on a dry tissue paper to gently remove any excess TBS buffer.
Transfer the membrane to a dry container and add the substrate to cover the entire membrane. Incubate 5 minutes with gentle rocking, at room temperature.
Remove the excess substrate by gently blotting the membrane on tissue paper.
Expose the membrane to a chemiluminescence film. The length of the exposure will need to be optimized to avoid signal saturation but will typically range from 10 seconds to 10 minutes.
Perform densitometry analyses on the developed film using a gel imaging apparatus such as the Fluorchem gel documentation system (Alpha Innotech) following the manufacturer’s instructions. The density values for each set of replicates of reference antigens and test vaccine samples are then averaged. While the antigen standards and vaccine samples can be run in duplicates, triplicates are preferred. Density values for replicates should be very similar. Significant differences between replicates indicate a probable technical problem during the procedure. A concentration-dependant standard curve is established by plotting the averaged density values versus the amount (in ng) of HA or NA antigen put in each slot, from the reference standard. Using appropriate calibration curve fitting in immunoassays is critical for quantifying HA and NA accurately. Linear regression, obtained by plotting the response (y) versus the concentration (x) using the linear portion of the response curve (y = mx + b) is the simplest method for the quantification of the analytes. If a broader range of analytes is considered for concentration calculation, the four parameter logistic (4PL) model for immunoassays currently accepted should be used. A variety of software is available for interested investigators to consider. In the case of the described slot blot assay, converting the x-axis values to log scale allows the curve to fit a 4-PL model and the use of a variable slope non-linear regression to calculate the blotted amounts of HA and NA in the tested vaccine samples is thus possible. We therefore routinely use this model for our analyses. As the test vaccine samples were diluted in either 4M urea/TBS or 0.01% Zwittergent/TBS prior to being blotted on the membrane, the dilution factor for each sample needs to be taken into account to determine the HA and NA content of the original test vaccine samples. It is suggested that each test vaccine sample be run at three different dilutions (2-fold difference) in order for one set of density values to fall within the standard curve. Density values higher than the standard curve range for all tested dilutions of the vaccine samples need to be further diluted for accurate HA or NA quantification. If density values are below the lowest end of the curve, test samples should be diluted at a lower dilution factor.
6. Representative Results:
Bioinformatics analyses of all available influenza A HA sequences confirmed the N-terminus of the HA2 subunit (the fusion peptide) as the only conserved region of HA. Figure 1 shows the conservation rate for each amino acid position of the identified consensus sequence. Two variations were identified at positions 2 (L—>I) and 12 (G—>N) of the 14 N-terminal amino acids of HA2, but such variations were found not to affect the binding between the antibodies and the peptide variants6. The Uni-1 epitope (GLFGAIAGFIEGGW) was chosen to develop a universal antibody against HA. This antibody demonstrated remarkable specificity for viral sequences and is capable of binding to 13 different subtypes of influenza A HA (H1-H13) (Figure 2).
Using a similar approach, two universally conserved sequences were identified in all influenza A NA’s, one close to the enzymatically active site (HCA-2) (Figure 3A) and the other at the N-terminus (HCA-3) (Figure 3B). Peptides with these amino acid sequences were used as antigens to generate universal antibodies against NA. The antibodies against both epitopes were capable of binding to all 9 subtypes of NA and showed very little cross-reactivity to allantoic or cellular proteins, thus demonstrating high specificity for viral NA sequences (Figure 4).
Figure 5 shows an overview of the slot blot method for the quantification of influenza HA and NA.
Examples of HA and NA antigen slot blot analysis are shown in Figures 6 and 7. Figure 6A and 6C shows representative results after the detection of the HA antigen in an influenza vaccine reference standard and vaccine samples respectively. Each sample (standard or vaccine) was run in duplicate, in adjacent wells. Duplicates should show similar intensities following detection. Optimization of antibody dilutions, incubation and exposure time may be required depending on the antibodies used.
An example of a typical standard curve obtained following the detection of various concentrations of HA from a vaccine reference standard sample is shown in Figure 6B. The concentration of HA antigen is proportional to the signal intensity following chemiluminescence detection of the slot blot and fits the 4-PL curve fitting model for this concentration range (0.0938 to 6 μg HA/ml).
Figure 7 shows representative results of the detection of the NA antigen in an influenza vaccine reference standard and in vaccine samples. Density analysis showed that the intensity of the signal obtained by detecting the NA antigen in various dilutions of a vaccine reference standard by slot blot is proportional to the concentration of NA (Fig. 7A). The resulting standard curve fits a 4-PL model for this concentration range (0.039 to 10 μg NA/ml), as shown in Figure 7B. Figure 7C shows an example of a slot blot analysis of the NA content in influenza vaccine samples. Each sample was assayed in duplicate, in adjacent wells.
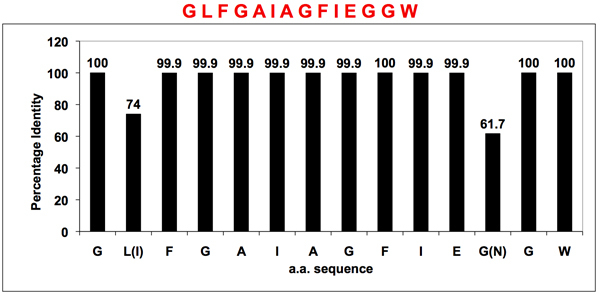 Figure 1. Conservation rate of the Uni-1 epitope in the fusion peptide region of influenza A HA.
Bioinformatics analyses of all available influenza A HA sequences confirmed that the fusion peptide (located at the N-terminus of the HA2 subunit) is the only universally conserved region of HA. Note that the two variations at position 2 (L—>I) and 12 (G—>N) were found not to affect the antibody binding6. A universal antibody capable of binding 13 different subtypes of influenza A HA (H1-H13) was developed using the Uni-1 epitope (GLFGAIAGFIEGGW).
Figure 1. Conservation rate of the Uni-1 epitope in the fusion peptide region of influenza A HA.
Bioinformatics analyses of all available influenza A HA sequences confirmed that the fusion peptide (located at the N-terminus of the HA2 subunit) is the only universally conserved region of HA. Note that the two variations at position 2 (L—>I) and 12 (G—>N) were found not to affect the antibody binding6. A universal antibody capable of binding 13 different subtypes of influenza A HA (H1-H13) was developed using the Uni-1 epitope (GLFGAIAGFIEGGW).
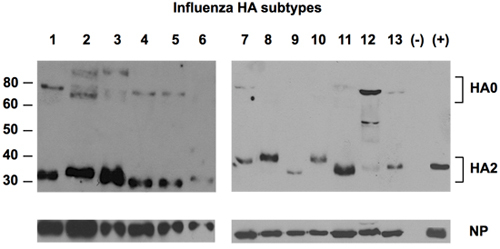 Figure 2. Detection of 13 subtypes of HA by universal antibody against HA.
Allantoic fluids of 13 subtypes of influenza A viruses propagated in embryonated eggs were fractionated in SDS-PAGE, followed by detection of the HA proteins using the universal antibody against the Uni-1 epitope of HA. NP protein was detected as loading control using a rabbit polyclonal NP-specific antibody. Negative control (-) was allantoic fluid from uninfected eggs. Positive control (+) was a reference standard of H1 from NIBSC, U.K.
Figure 2. Detection of 13 subtypes of HA by universal antibody against HA.
Allantoic fluids of 13 subtypes of influenza A viruses propagated in embryonated eggs were fractionated in SDS-PAGE, followed by detection of the HA proteins using the universal antibody against the Uni-1 epitope of HA. NP protein was detected as loading control using a rabbit polyclonal NP-specific antibody. Negative control (-) was allantoic fluid from uninfected eggs. Positive control (+) was a reference standard of H1 from NIBSC, U.K.
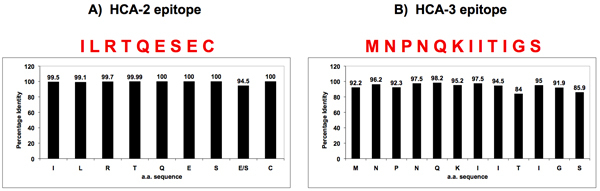 Figure 3. Sequence homology of influenza A NA’s near the enzymatically active site and the N-terminus.
Two universally conserved sequences were identified in influenza A NA’s by bioinformatics, one located near the enzymatic active site (HCA-2) (Panel A), the other at the N-terminus (HCA-3) (Panel B). Peptides with these conserved amino acid sequences were synthesized and used to generate universal antibodies against NA.
Figure 3. Sequence homology of influenza A NA’s near the enzymatically active site and the N-terminus.
Two universally conserved sequences were identified in influenza A NA’s by bioinformatics, one located near the enzymatic active site (HCA-2) (Panel A), the other at the N-terminus (HCA-3) (Panel B). Peptides with these conserved amino acid sequences were synthesized and used to generate universal antibodies against NA.
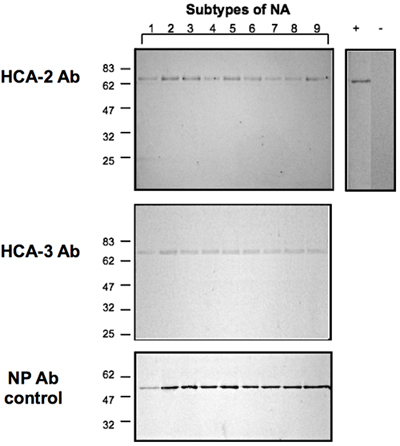 Figure 4. Detection of 9 subtypes of NA by NA antibodies.
Allantoic fluids of nine NA subtypes of influenza A viruses propagated in embryonated eggs were fractionated in SDS-PAGE, followed by detection of the NA proteins using the HCA-2 (Panel A) and HCA-3 (Panel B) antibodies. NP protein was detected as loading control using a rabbit polyclonal NP-specific antibody (Panel C). Negative control (-) was allantoic fluid from uninfected eggs. Positive control (+) was allantoic fluid spiked with rNA1 of A/New Caledonia/20/99 and probed with the corresponding antisera.
Figure 4. Detection of 9 subtypes of NA by NA antibodies.
Allantoic fluids of nine NA subtypes of influenza A viruses propagated in embryonated eggs were fractionated in SDS-PAGE, followed by detection of the NA proteins using the HCA-2 (Panel A) and HCA-3 (Panel B) antibodies. NP protein was detected as loading control using a rabbit polyclonal NP-specific antibody (Panel C). Negative control (-) was allantoic fluid from uninfected eggs. Positive control (+) was allantoic fluid spiked with rNA1 of A/New Caledonia/20/99 and probed with the corresponding antisera.
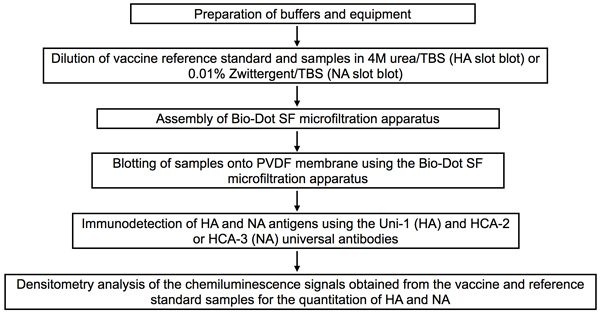 Figure 5. Flow chart of the slot blot procedure for the quantitation of HA and NA antigens in influenza vaccines.
The buffers needed for sample dilution as well as for washing and blocking the slot blot membrane are first prepared and the PVDF membrane and filter papers needed for assembly of the Bio-Dot SF microfiltration apparatus are pre-soaked. Influenza vaccine and reference standard samples are diluted in the optimal buffers for HA or NA detection by slot blot. The Bio-Dot SF microfiltration apparatus is assembled according to manufacturer’s instructions and samples are applied onto the PVDF membrane. HA and NA antigens are detected using the universal antibodies against each antigen. Densitometry analysis is performed on the chemiluminescence signals obtained for quantitation of HA and NA in the tested samples.
Figure 5. Flow chart of the slot blot procedure for the quantitation of HA and NA antigens in influenza vaccines.
The buffers needed for sample dilution as well as for washing and blocking the slot blot membrane are first prepared and the PVDF membrane and filter papers needed for assembly of the Bio-Dot SF microfiltration apparatus are pre-soaked. Influenza vaccine and reference standard samples are diluted in the optimal buffers for HA or NA detection by slot blot. The Bio-Dot SF microfiltration apparatus is assembled according to manufacturer’s instructions and samples are applied onto the PVDF membrane. HA and NA antigens are detected using the universal antibodies against each antigen. Densitometry analysis is performed on the chemiluminescence signals obtained for quantitation of HA and NA in the tested samples.
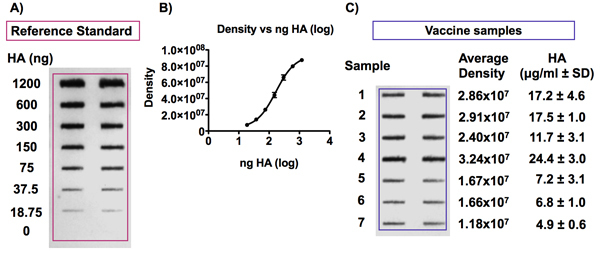 Figure 6. Detection of the HA antigen in influenza vaccine samples and reference standard.
Panel A shows a representative blot obtained following immunodetection of the HA antigen. The HA reference antigen was diluted to concentrations from 0 to 6 μg HA/ml and the vaccines were diluted to a concentration of 0.375 μg HA/ml in 4M urea/TBS before being applied to a PVDF membrane with a Bio-Dot SF microfiltration apparatus. The HA antigen was then detected using the Uni-1 universal antibody. Panel B shows an example of a standard curve obtained by detecting signals of various HA concentrations from a vaccine reference standard. The signal intensity is proportional to the concentration of HA and the curve fits a 4-PL model.
Figure 6. Detection of the HA antigen in influenza vaccine samples and reference standard.
Panel A shows a representative blot obtained following immunodetection of the HA antigen. The HA reference antigen was diluted to concentrations from 0 to 6 μg HA/ml and the vaccines were diluted to a concentration of 0.375 μg HA/ml in 4M urea/TBS before being applied to a PVDF membrane with a Bio-Dot SF microfiltration apparatus. The HA antigen was then detected using the Uni-1 universal antibody. Panel B shows an example of a standard curve obtained by detecting signals of various HA concentrations from a vaccine reference standard. The signal intensity is proportional to the concentration of HA and the curve fits a 4-PL model.
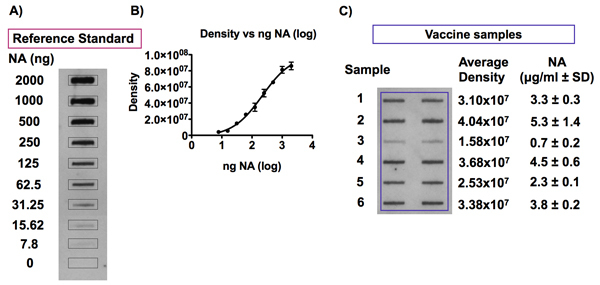 Figure 7. Detection of the NA antigen in influenza vaccine samples and reference standard.
Panel A and B show representative signals obtained following the detection of NA in an influenza vaccine reference standard diluted to NA concentrations ranging from 0 to 10 μg NA/ml in 0.01% Zwittergent/TBS and the resulting 4-PL standard curve following densitometry analyses. An example of typical chemiluminescence signals detected following a slot blot analysis of NA antigen in influenza vaccine samples is shown in Panel C. Influenza vaccine samples were diluted to a concentration of 1 μg HA/ml in 0.01% Zwittergent/TBS and were blotted onto a PVDF membrane using a Bio-Dot SF microfiltration apparatus. Dilutions of the corresponding vaccine reference standard, with concentrations ranging from 0 to 2.5 μg NA/ml, were included on the same blot in order to establish a standard curve for quantitation of the NA antigen in the vaccine samples. The NA antigen was detected with the HCA-2 universal antibody against NA and densitometry analysis of the chemiluminescence signals obtained was performed. The amount of NA in each vaccine sample can be calculated against the 4-PL standard curve obtained by plotting the signal intensity values versus the NA concentration for the vaccine reference standard.
Figure 7. Detection of the NA antigen in influenza vaccine samples and reference standard.
Panel A and B show representative signals obtained following the detection of NA in an influenza vaccine reference standard diluted to NA concentrations ranging from 0 to 10 μg NA/ml in 0.01% Zwittergent/TBS and the resulting 4-PL standard curve following densitometry analyses. An example of typical chemiluminescence signals detected following a slot blot analysis of NA antigen in influenza vaccine samples is shown in Panel C. Influenza vaccine samples were diluted to a concentration of 1 μg HA/ml in 0.01% Zwittergent/TBS and were blotted onto a PVDF membrane using a Bio-Dot SF microfiltration apparatus. Dilutions of the corresponding vaccine reference standard, with concentrations ranging from 0 to 2.5 μg NA/ml, were included on the same blot in order to establish a standard curve for quantitation of the NA antigen in the vaccine samples. The NA antigen was detected with the HCA-2 universal antibody against NA and densitometry analysis of the chemiluminescence signals obtained was performed. The amount of NA in each vaccine sample can be calculated against the 4-PL standard curve obtained by plotting the signal intensity values versus the NA concentration for the vaccine reference standard.
Discussion
Quantitative determination of influenza viral HA and NA are critical for vaccine research and development since these two surface proteins are most important viral components inducing immune responses6-11. Previously reported immunological methods for the detection of these proteins require strain specific antibodies. The simple, reproducible and rapid slot blot method to quantify the HA and NA antigens described here are suitable for all influenza A viral HA and NA proteins since the antibodies recognize their only universally conserved epitopes6-8. The sole difficulty for investigators interested in this method might be that they have no such universal antibodies in-house. However, generation of these antibodies using peptides as antigens is a routine procedure. While interested scientists can make the antibodies themselves according to the procedures described because the variants peptides compete equally well among themselves for binding to the given peptide in a competitive ELISA6-8, they may also contact the authors for the HA and NA antibodies to be used in their exploratory research.
As far as the slot blot assay is concerned, it is rather straightforward, with special attention needed at only a few steps, as described above. Although HA and NA could both be detected in untreated samples, increased sensitivity was achieved by a pre-treatment of samples.
Despite their advantages of simplicity, rapidity and reproducibility, the universal antibody-based slot blot methods described here have certain limitations. For example, they are not designed to distinguish different HA or NA subtypes and as such, are possibly more suitable for in-process quality control and/or analyses of monovalent vaccine preparations. The SRID assay therefore remains the method of choice to identify subtypes of viral HA in influenza vaccines3.
In short, the highly conserved sequences in HA and NA have been selected to generate mono-specific antibodies, which can be employed for quantitative analyses of virtually any strains of influenza viral HA and NA. While the detailed slot blot procedure with these universal antibodies against HA and NA is presented here, interested investigators could use them as a basic research tool in other types of immunoassay, given that they demonstrated remarkable specificity against viral sequences without detectable cross-reactivity with allantoic or cellular proteins. Also of note is that the universal antibodies against HA can detect HA in native conformation in virus-infected cell cultures12, which highlights a broader range of applications of these antibodies as basic research tools.
Disclosures
No conflicts of interest declared.
Acknowledgments
The authors would like to thank Mrs. Monika Tocchi for editorial review of the manuscript. AMH is supported by a Scholarship from King Abdulaziz University, through the Saudi Arabian Cultural Bureau in Canada.
References
- Webster RG, Bean WJ. Genetics of influenza virus. Annu Rev Genet. 1978;12:415–431. doi: 10.1146/annurev.ge.12.120178.002215. [DOI] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Wood JM. The influence of the host cell on standardisation of influenza vaccine potency. Dev Biol Stand. 1999;98:183–188. [PubMed] [Google Scholar]
- Sylte MJ, Suarez DL. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol. 2009;333:227–241. doi: 10.1007/978-3-540-92165-3_12. [DOI] [PubMed] [Google Scholar]
- Bright RA, Neuzil KM, Pervikov Y, Palkonyay L. WHO meeting on the role of neuraminidase in inducing protective immunity against influenza infection. Vaccine. 2008;27:6366–6369. doi: 10.1016/j.vaccine.2009.02.084. [DOI] [PubMed] [Google Scholar]
- Chun S. Universal antibodies and their applications to the quantitative determination of virtually all subtypes of the influenza A viral hemagglutinins. Vaccine. 2008;26:6068–6076. doi: 10.1016/j.vaccine.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Gravel C. Qualitative and quantitative analyses of virtually all subtypes of influenza A and B viral neuraminidases using antibodies targeting the universally conserved sequences. Vaccine. 2010;28:5774–5784. doi: 10.1016/j.vaccine.2010.06.075. [DOI] [PubMed] [Google Scholar]
- Li C. A simple slot blot for the detection of virtually all subtypes of the influenza A viral hemagglutinins using universal antibodies targeting the fusion peptide. Nat Protoc. 2010;5:14–19. doi: 10.1038/nprot.2009.200. [DOI] [PubMed] [Google Scholar]
- Harvey R, Wheeler JX, Wallis CL, Robertson JS, Engelhardt OG. Quantitation of haemagglutinin in H5N1 influenza viruses reveals low haemagglutinin content of vaccine virus NIBRG-14 (H5N1) Vaccine. 2008;26:6550–6554. doi: 10.1016/j.vaccine.2008.09.050. [DOI] [PubMed] [Google Scholar]
- Li C. Application of deglycosylation and electrophoresis to the quantification of influenza viral hemagglutinins facilitating the production of 2009 pandemic influenza (H1N1) vaccines at multiple manufacturing sites in China. Biologicals. 2010;38:284–289. doi: 10.1016/j.biologicals.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Johansson BE, Pokorny BA, Tiso VA. Supplementation of conventional trivalent influenza vaccine with purified viral N1 and N2 neuraminidases induces a balanced immune response without antigenic competition. Vaccine. 2002;20:1670–1674. doi: 10.1016/s0264-410x(01)00490-x. [DOI] [PubMed] [Google Scholar]
- Hashem A. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochem Biophys Res Comm. 2010 doi: 10.1016/j.bbrc.2010.11.030. Forthcoming. [DOI] [PubMed] [Google Scholar]


