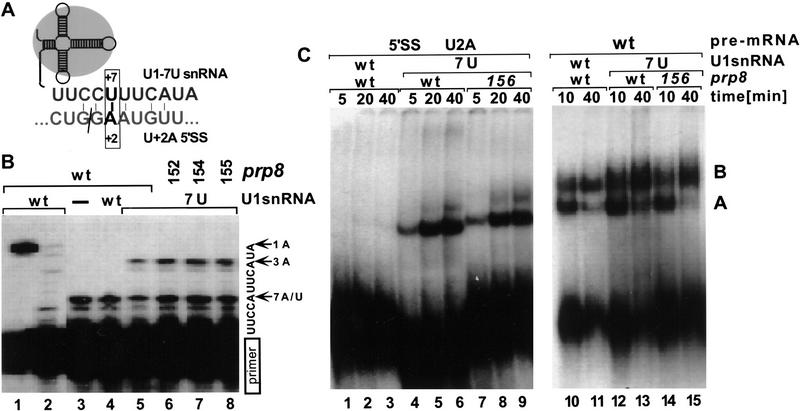
Figure 4.
Suppressor effects of U1–7U snRNA. (A) Schematic representation of the U+2A 5′SS and U1–7U snRNA pairing interaction. (B) Primer extension analysis of total RNA isolated from yeast extracts containing wild-type (lanes 1–5) or suppressor prp8-152, prp8-154, and prp8-155 (lanes 6–8). The analyzed strains contained in addition a wild-type U1 (lanes 1,2,4) or suppressor U1–7U (lanes 5–8) snRNA gene. To detect both the wild-type and U1–7U snRNAs, primer extension analysis was carried out in the presence of all dNTPs (lane 1), dGTP and dATP (lane 2) or dGTP, dATP, ddTTP (lanes 3–8). In the presence of ddTTP, the 5-nucleotide extension corresponds to a wild-type snRNA, and a 9-nucleotide extension to a U1–7U snRNA. (C) In vitro splicing reactions using U+2A (lanes 1–9) and wild-type (lanes 10–15) pre-mRNA were carried out in the presence of extracts prepared from the wild-type PRP8 (lanes 1–6,10–13) or prp8-156 strain (lanes 7–9,14,15) containing wild-type (lanes 1–3,10,11) or U1–7U snRNA (lanes 4–9,12–15). Aliquots withdrawn at times indicated were resolved in a native gel. Positions of prespliceosome (A) and spliceosome (B) are indicated.