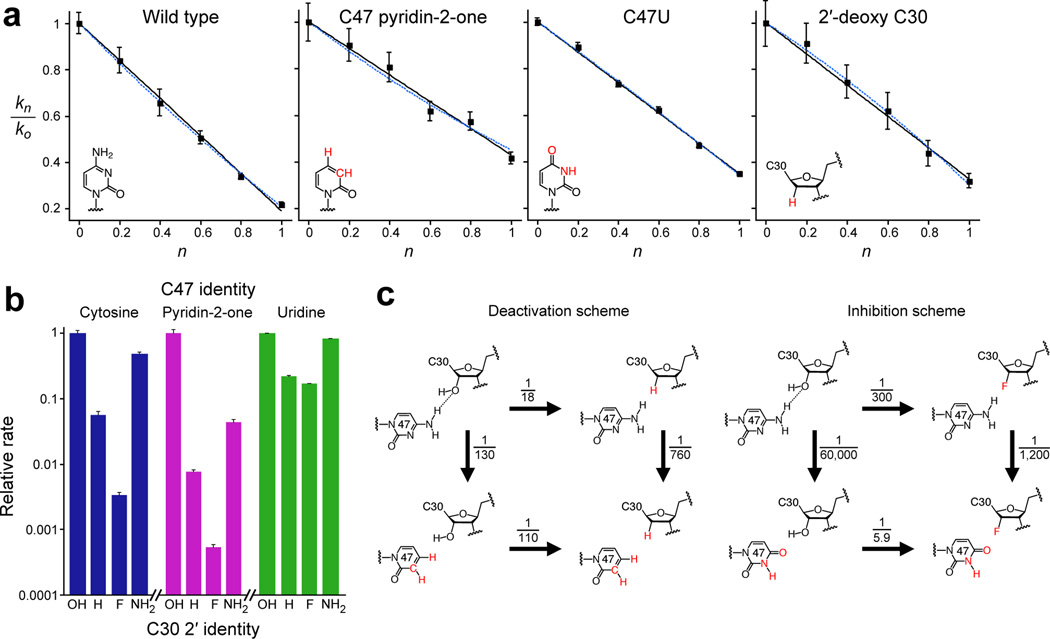
Figure 4.
Catalytic roles of active-site functional groups. a. Proton inventories for self-ligation by the indicated variants. The solid black line indicates the best fit for a single-proton–transfer model; the dashed blue line indicates the best fit for a two-proton–transfer model28,37,38. Error bars, standard deviation from three independent experiments. b. Relative rates for self-ligation of constructs with the indicated substitutions at the 2′ position of C30, surveyed in combination with the indicated subsitutions at position 47. For each C47 substitution, rates are normalized to that of the C30 2′-OH construct. Error bars, standard deviation from three independent experiments. c. Schemes summarizing functional interaction between the C30 2′-hydroxyl and the C47 N4, showing the effects of removing these groups (left), or replacing them with inhibitory groups (right). Substitutions, relative to the wild type, denoted in red. Numbers indicate the decrease in self-ligation rate, (kright/kleft) or (kbottom/ktop), resulting from the indicated substitution. At pH 6.0, the observed rate of the unmodified ligase (upper left corner of each scheme) was 0.93 ± 0.009 min−1 (s.d.).