Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Resolution (Å) | 50.0–2.68 (2.75–2.68) |
| Wavelength (Å) | 0.9791 |
| Beamline | 24-ID-C, APS |
| Space group | P212121 |
| Unit-cell parameters | |
| a (Å) | 59.7 |
| b (Å) | 140.1 |
| c (Å) | 143.3 |
| No. of reflections | 87487 |
| Unique reflections | 31670 |
| Average I/σ(I) | 12.2 (1.5) |
| Multiplicity | 2.8 (1.5) |
| Completeness (%) | 92.8 (70.0) |
| Rmerge† (%) | 9.4 (32.9) |
| Data refinement | |
| No. of protein atoms | 7795 |
| No. of ligand atoms | 5 |
| No. of water atoms | 56 |
| Working-set reflections | 30019 |
| Test-set reflections | 1606 |
| R factor‡ | 23.8 |
| Rfree§ | 27.8 |
| R.m.s.d. bonds (Å) | 0.006 |
| R.m.s.d. angles (°) | 0.91 |
| Mean B factor (Å2) | 53 |
| Ramachandran plot | |
| Most favored (%) | 92.7 |
| Additionally allowed (%) | 7.1 |
| Generously allowed (%) | 0.1 |
| Disallowed (%) | 0.1¶ |
R
merge = 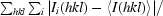
 , where 〈I(hkl)〉 is the mean intensity of the i reflections with intensities I
i(hkl) and common indices hkl.
, where 〈I(hkl)〉 is the mean intensity of the i reflections with intensities I
i(hkl) and common indices hkl.
R factor = 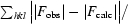
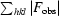 , where F
obs and F
calc are observed and calculated structure factors, respectively.
, where F
obs and F
calc are observed and calculated structure factors, respectively.
For R free, the sum is extended over a subset of reflections (5%) that were excluded from all stages of refinement.
Disallowed angles occurred at a Gly-Gly sequence (residues 200–201).
