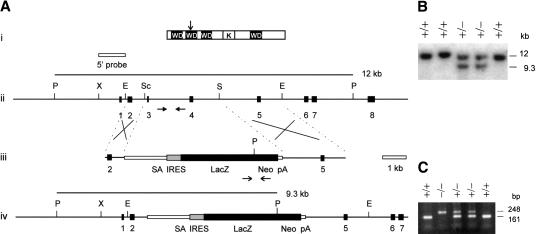
Figure 1.
Targeted disruption of the mouse Bub3 gene. (A) Gene disruption construct and restriction maps. (i) Mouse Bub3 protein showing positions of four WD40 motifs (WD) and the Bub1-kinetocore-interacting domain (K) (Taylor et al. 1998; Martinez-Exposito et al. 1999). The disruption site is indicated by the vertical arrow. Restriction-enzyme maps for (ii) the Bub3 gene covering exons 1 to 8, (iii) the neomycin-resistance gene targeting construct, and (iv) the Bub3 locus following targeted disruption. Black boxes represent exons. The selectable marker cassette contained in the targeting construct consists of a splice acceptor site (SA), a picornaviral internal ribosome-entry site (IRES), a lacZ–neomycin fusion gene (beta-geo) and a SV40 polyadenylation sequence (pA). A 1.2-kb XbaI–EagI fragment (designated 5′ probe) spanning exon 1 was used in the Southern-screening strategy and detected a 12-kb wild-type PstI fragment in the untargeted locus or a 9.3-kb PstI fragment in the targeted locus. The positions of the primers for nested PCR genotyping of cultured embryos up to day 3.5 + 4 are shown by the horizontal arrows in (ii) and (iii). Wild type primer pairs include BI3H-BI3I for the first round and BI3J-BI3K for the second round of amplification (closed horizontal arrows in ii), giving a final product of 161 bp for the untargeted allele. Beta-geo primer pairs include GF1–GR1 for first-round synthesis and GF2–GR2 for second-round synthesis (open horizontal arrows in iii), giving a final product of 248 bp for the targeted allele. Crosses denote expected sites of homologous recombination. Abbreviations for restriction enzymes are (E) EagI, (P) PstI, (Sc) SacI, (S) SalI, and (X) XbaI. (B) Southern blot analysis of wild-type and gene-targeted ES cell lines. The sizes of wild-type 12-kb and homologous recombinant 9.3-kb bands are shown. (C) Nested PCR analysis of cultured 3.5 + 4 day embryos from heterozygous crosses showing the expected bands for the targeted (248 bp) and untargeted (161 bp) alleles.