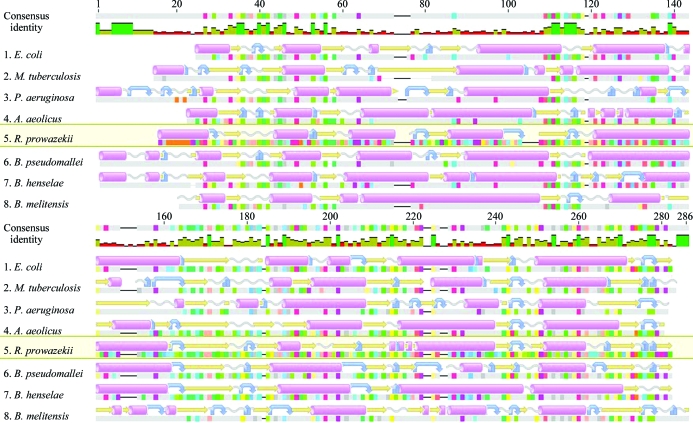
Figure 4.
Alignment of solved structures of bacterial 3-ketoacyl-(ACP) reductases. Multiple sequence and secondary-structure alignment of bacterial orthologs. Numbering along the top is based on the alignment consensus. Predicted α-helices, β-strands, coils and turns are shown as pink cylinders, yellow arrows, grey corkscrews and blue curved arrows, respectively. Sequences are taken from the PDB. From top to bottom: E. coli, 1i01 (Price et al., 2001 ▶); M. tuberculosis, 1uzm (Cohen-Gonsaud et al., 2002 ▶); Pseudomonas aeruginosa, 2b4q (Miller et al., 2006 ▶); Aquifex aeolicus, 2pnf (Q. Mao, R. Huether, W. L. Duax & T. C. Umland, unpublished work); R. prowazekii, 3f9i (this work); Burkholderia pseudomallei, 3ftp (Seattle Structural Genomics Center for Infectious Disease, unpublished work); Bartonella henselae, 3grp (B. L. Staker, unpublished work); Brucella melitensis, 3n74 (Seattle Structural Genomics Center for Infectious Disease, unpublished work). This figure was prepared with Geneious (Drummond et al., 2010 ▶).