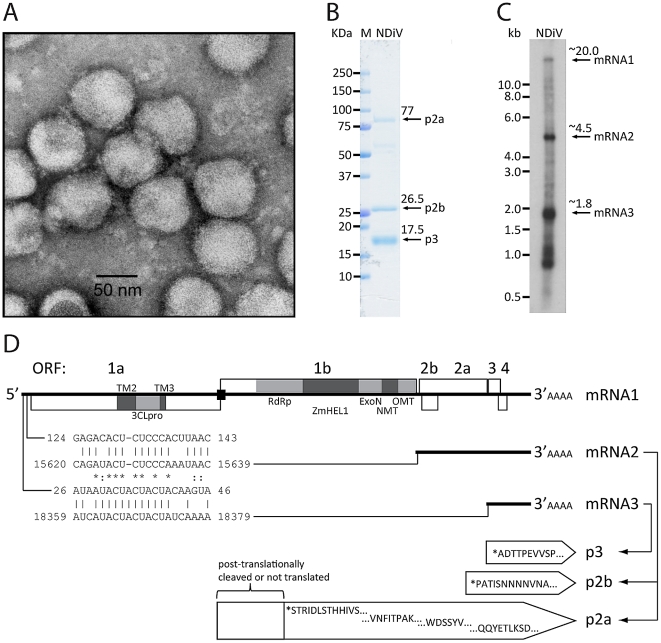
Figure 3. NDiV characteristics.
(A) Electron micrograph of negatively stained NDiV virions. (B) SDS-PAGE analysis of NDiV virion proteins. (C) Detection of NDiV genomic (mRNA1) and subgenomic mRNAs (mRNAs 2 and 3) by Northern blot hybridization analysis of total intracellular RNA from virus-infected cells, using a radiolabeled probe complementary to the 3′end of the NDiV genome. (D) NDiV genome organization and expression. Open reading frames (ORFs) are represented by open rectangles and ORF1a- and ORF1b-encoded protein domains identified by bioinformatics analyses (see Table 2) are highlighted in grey. Peptide sequences of virion proteins were determined as described in the Materials and Methods section and mapped to the products of ORFs 2a, 2b, and 3 (bottom-right). N-terminal protein sequences are indicated by (*), other peptide sequences indicate inner sequences. The actual molecular size of the ORF2a product (approximately 77 KDa in SDS-PAGE in B) is considerably shorter than the calculated size (102 KDa), suggesting that p2a may be post-translationally proteolytically processed or that its translation starts at another AUG codn in the ORF. Two pairs of conserved potential TRSs – for sg mRNAs 2 and 3, respectively - were identified in the NDiV genome and aligned (bottom-left), with each pair consisting of a putative leader TRS in the 5′-UTR and a body TRS in the 3′-proximal region of the genome. Between these TRS pairs, eight and three positions include complete match (*) and nucleotide overlap (:), respectively.