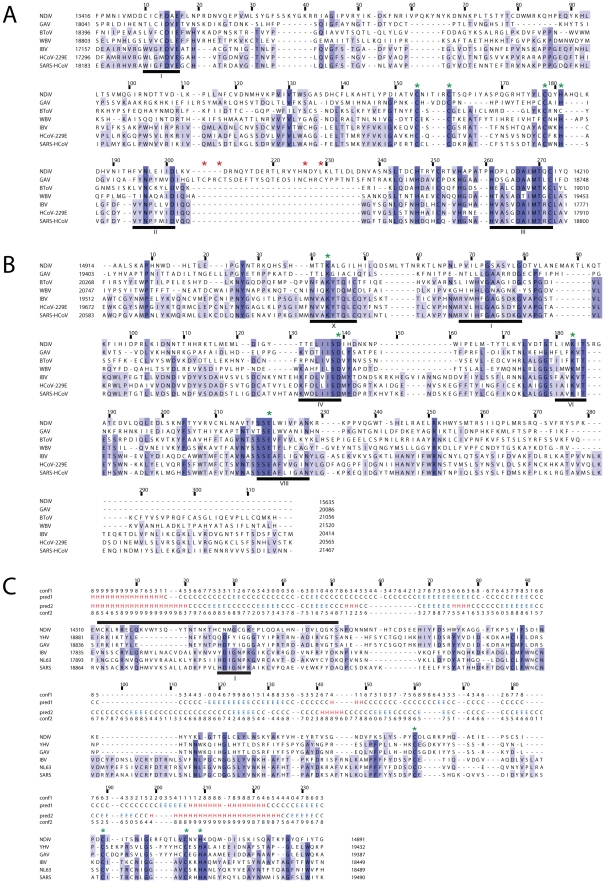
Figure 5. Alignments of ExoN, OMT and NMT domains of NDiV and other nidoviruses.
Alignments were compiled utilizing the Muscle program followed by manual inspection. Pictures by JalView [90] and residues are colored according to degree of conservation. Numbers above a column indicate its absolute position in the alignment (start = 1); numbers to the left and to the right of the alignment represent positions in the genome. Selected conserved sequence motifs are highlighted with black bars and roman numbers. (A) In the exonuclease (ExoN) alignment, three motifs are part of the catalytic centre; the domain includes two putative zinc fingers, specific either for roniviruses or for all nidoviruses and highlighted by, respectively, red and green asterisks. (B) In the 2′-O-methyltransferase (OMT) alignment, motifs X, IV, VI and VIII include residues of the catalytic tetrad (KDKE, marked with green asterisks) and motif I is involved in binding of the methyl donor [42]. (C) Protein secondary structure predictions by Psipred [86] for the profiles of N7-methyltransferase (NMT) from 3 NDiV/roniviruses (pred1) and 17 coronaviruses (pred2) and corresponding confidence values (conf1, conf2) were added above the alignment. Only 3 coronaviruses, representing alpha- (HCoV NL63), beta- (SARS-CoV) and gammacoronaviruses (IBV), are shown that results in several empty alignment columns. The black bar on top is a region including the methyl-donor binding site (motif I, delineated by [49]) that gave a hit with a functionally similar site of a cellular guanine N7-methyltransferase (fungus Encephalitozoon cuniculi) upon HH search of the SCOP database [82] (data not shown). Green asterisks, conserved Cys/His residues that may form a zinc finger.