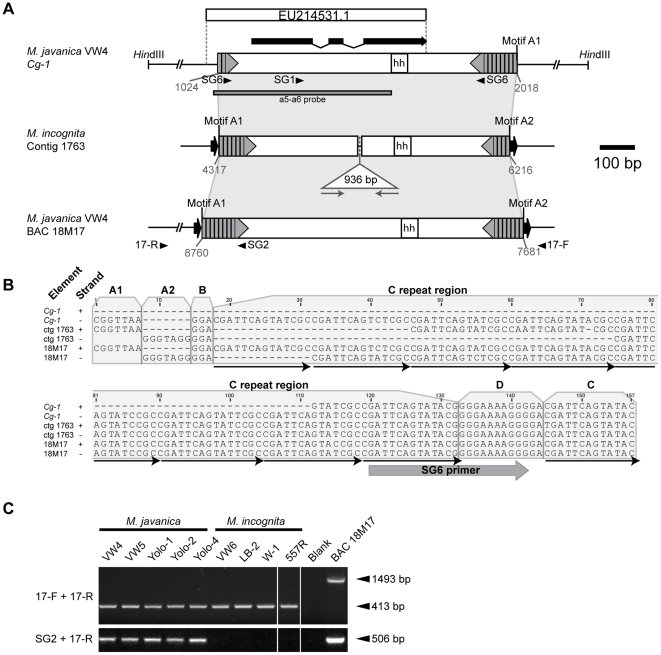
Figure 1. Inverted-repeat element carrying Cg-1 and related elements.
(A) Alignment between elements (open boxes) at the M. javanica VW4 Cg-1 locus (GenBank EU214531.2), M. incognita contig 1763 (CABB01001763.1), and M. javanica VW4 BAC 18M17 (HQ122410.1). Similar regions between elements are designated with light grey shading. Position of Cg-1 transcript [24] is shown with thick black lines representing exons and thin lines, introns. Dark grey arrows within open boxes represent the TIRs with vertical bars indicating internal repeats. Target site duplications are indicated black arrows; hh designates the position of the histone hairpin. Binding sites for primers SG2, SG6, 17-F and 17-R are indicated by arrowheads. The grey bar (a5–a6 probe) shows the position of the PCR amplicon used for Southern analysis. (B) Alignment of TIRs of the elements shown in (A). Strand designations+and−represent the 5′ and 3′ ends for the elements as drawn above. Motifs and binding site of primer SG6 are indicated. Thin black arrows below the alignment denote repeat units of Motif C. (C) Agarose gel showing amplification products from genomic DNA of M. incognita and M. javanica isolates using primers 17-F and 17-R for amplification of empty sites and primer set SG2 and 17-R to detect an insertion similar to that on BAC 18M17.