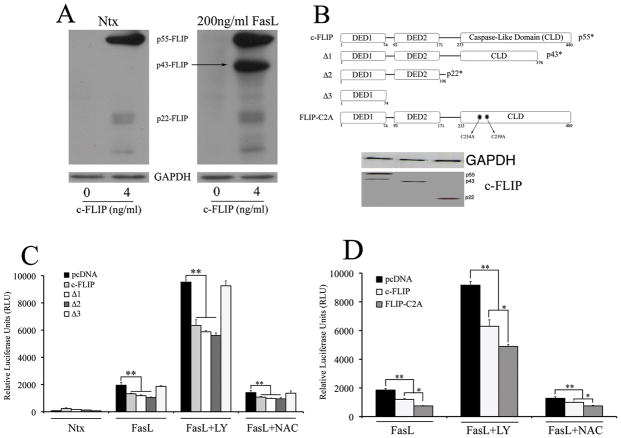
Fig. 7. Role of c-FLIP processing and S-nitrosylation in FasL-induced NF-κB activation.
A. Cells transfected with either pcDNA3 control plasmid or 4ng/ml c-FLIP were exposed to 200ng/ml FasL, and probed for c-FLIP levels using western blotting. Blots were re-probed with GAPDH antibody to confirm equal loading of the samples. B. Domain mutants were generated using standard techniques as mentioned in the material and methods section and were cloned into the pcDNA3 vector. The physiologically relevant processed forms include p43-FLIP (D1), p22-FLIP (D2) and domain mutant expressing only the death effector domain 1 (D3). In addition, full-length c-FLIP with the two cysteines in its caspase-like domain substituted by alanines (D4) was generated such that the mutated form cannot undergo S-nitrosylation. Western blots probed with c-myc antibody show the expression of the c-FLIP domain mutants. C. Cells co-transfected with 100ng/well NF-κB-Luc, 10ng/well pRL-tk and domain mutants of c-FLIP (4ng/ml) were treated with 200ng/ml FasL (12 h) in the absence or presence of 20μM LY294002 and 10mM NAC (1 h pre-treatment), and then assayed for NF-κB activity by luciferase assay. D. Cells were co-transfected with 100ng/well NF-κB-Luc, 10ng/well pRL-tk and non-nitrosylable c-FLIP mutant (D4). Transfected cells were either left untreated or pretreated with 20μM LY294002 and 10mM NAC for 1 h followed by 200ng/ml FasL treatment for 12 h. Treated cells were analyzed for NF-κB activity by luciferase assay. Values are mean ± SD (n = 4). *P < 0.05, **P < 0.05 for respective data versus pcDNA-transfected control datasets as indicated.