Abstract
Translation initiation factor 2 (eIF2) bound to GTP transfers the initiator methionyl tRNA to the 40S ribosomal subunit. The eIF5 stimulates GTP hydrolysis by the eIF2/GTP/Met-tRNAiMet ternary complex on base-pairing between Met-tRNAiMet and the start codon. The eIF2, eIF5, and eIF1 all have been implicated in stringent selection of AUG as the start codon. The eIF3 binds to the 40S ribosome and promotes recruitment of the ternary complex; however, physical contact between eIF3 and eIF2 has not been observed. We show that yeast eIF5 can bridge interaction in vitro between eIF3 and eIF2 by binding simultaneously to the amino terminus of eIF3 subunit NIP1 and the amino-terminal half of eIF2β, dependent on a conserved bipartite motif in the carboxyl terminus of eIF5. Additionally, the amino terminus of NIP1 can bind concurrently to eIF5 and eIF1. These findings suggest the occurrence of an eIF3/eIF1/eIF5/eIF2 multifactor complex, which was observed in cell extracts free of 40S ribosomes and found to contain stoichiometric amounts of tRNAiMet. The multifactor complex was disrupted by the tif5-7A mutation in the bipartite motif of eIF5. Importantly, the tif5-7A mutant is temperature sensitive and displayed a substantial reduction in translation initiation at the restrictive temperature. We propose that the multifactor complex is an important intermediate in translation initiation in vivo.
Keywords: Eukaryotic translation initiation factor (eIF), ribosomal preinitiation complex, GAP, AUG selection, Met-tRNAiMet binding
The process of translation initiation involves the formation of a ribosomal initiation complex in which the anticodon of methionyl initiator tRNA (Met-tRNAiMet) is base-paired with the start codon in the mRNA. In eukaryotes, AUG is selected with high stringency as the start codon, and in most cases, the AUG triplet closest to the 5′ end is used regardless of the structure of the mRNA. Numerous protein factors called eukaryotic initiation factors (eIFs) interact with the 40S ribosomal subunit to stimulate assembly of the translation initiation complex (for review, see Merrick and Hershey 1996). According to a widely accepted model, the large multisubunit factor called eIF3 binds first to the free 40S subunit, and impedes association with the 60S ribosomal subunit. The Met-tRNAiMet is then delivered to the 40S subunit in a ternary complex consisting of eIF2 (a heterotrimeric factor) bound to a molecule of GTP. The resulting 43S preinitiation complex interacts with an mRNA molecule containing eIF4F bound to the m7GpppN cap structure at the 5′ end. The mRNA-bound 48S complex facilitates the recognition of AUG triplets by the anticodon of Met-tRNAiMet in a process known as scanning. The correct codon–anticodon pairing stimulates hydrolysis of the GTP bound to eIF2 in a reaction that is stimulated by the GTPase activating protein (GAP) eIF5, and probably modulated by eIF1. Hydrolysis of GTP triggers the release of eIF2–GDP and other eIFs from the ribosome, producing a 40S initiation complex that can join with the 60S subunit. Recently, a new factor known as eIF5B (yIF2 in yeast; Choi et al. 1998) was found to interact with the 60S subunit and stimulate subunit joining in a reaction that consumes a second molecule of GTP (Pestova et al. 2000). The resulting 80S initiation complex, with Met-tRNAiMet bound to the P-site, is ready to accept the next incoming aminoacyl–tRNA in the A-site. Because only the GTP-bound form of eIF2 can bind Met-tRNAiMet, the GDP bound to eIF2 at the end of this process must be replaced by GTP in a reaction dependent on the heteropentameric guanine nucleotide exchange factor (GEF) eIF2B.
It was shown that eIF3 associates with the 40S ribosome and stimulates the binding of Met-tRNAiMet in partial reactions reconstituted in vitro with purified mammalian eIFs (Benne and Hershey 1978; Trachsel and Staehelin 1979). In accordance with these findings, a temperature sensitive (Ts−) yeast strain bearing the prt1-1 mutation in the 90 kD subunit of eIF3 (Naranda et al. 1994) was defective for Met-tRNAiMet binding to 40S ribosomes at the restrictive temperature (Feinberg et al. 1982), and this defect could be complemented in mutant cell extracts by purified eIF3 (Danaie et al. 1995; Phan et al. 1998). However, because there is no evidence for direct interaction between eIF3 and eIF2, the stimulatory effect of eIF3 on ternary complex binding might be mediated by other eIFs, or could involve allosteric alteration of the ribosome by eIF3.
In contrast to the paucity of evidence for eIF2–eIF3 physical association, there is strong evidence that eIF1 (known as SUI1) and eIF5 interact with yeast eIF3 (Naranda et al. 1996; Phan et al. 1998). The eIF3–eIF5 association has also been observed in the mammalian system (Bandyopadhyay and Maitra 1999). Interestingly, eIF1 and eIF5 both interacted with the NIP1 encoded 93-kD subunit of yeast eIF3 in vitro (Asano et al. 1998; Phan et al. 1998), and the same is true for the mammalian eIF1 and NIP1 homologs (Fletcher et al. 1999). Both eIF5 and eIF1, together with all three subunits of eIF2, have been implicated in accurate recognition of initiation codons in yeast (Donahue et al. 1988; Cigan et al. 1989; Castilho-Valavicius et al. 1990; Yoon and Donahue 1992; Dorris et al. 1995; Huang et al. 1997). It has been proposed that the stringency of AUG selection is determined by the propensity of eIF2 to hydrolyze GTP bound to the ternary complex, and the ability of eIF5 to stimulate this reaction during the scanning process (Huang et al. 1997). Mammalian eIF1 is required for formation of a 48S complex capable of locating the first AUG (Pestova et al. 1998), although its biochemical function is unknown. Based on the mutual association of eIF5 and eIF1 with eIF3–NIP1, we proposed that eIF3 may properly juxtapose these factors in relation to the ternary complex and mRNA on the 40S ribosome for accurate AUG selection (Phan et al. 1998).
The carboxy-terminal ∼40% of yeast eIF5 harbors the binding domain for eIF3–NIP1 and contains a bipartite sequence motif containing conserved aromatic and acidic residues (AA-boxes 1 and 2) that is required for interaction between eIF5 and both isolated NIP1 and purified eIF3 in vitro (Asano et al. 1999). It is intriguing that the carboxy-terminal segment of eIF5, including the AA-boxes, is also required for stable association of eIF5 with its substrate eIF2. The fact that the AA-box domain in eIF5 can interact with either eIF3–NIP1 or eIF2β, plus the observation that both eIF2 and eIF3 each were coimmunoprecipitated with eIF5 from yeast extracts (Asano et al. 1999), raised the possibility that a multi-eIF complex containing eIF3, eIF1, eIF5, and the eIF2/GTP/Met-tRNAiMet ternary complex may exist in the cytoplasm and bind to the 40S ribosome as a preformed unit.
We provide biochemical data indicating the existence of a multifactor eIF3/eIF1/eIF5/eIF2/GTP/Met-tRNAiMet complex that can exist free of the ribosome and is dependent for its integrity on the AA-box domain at the carboxyl terminus of eIF5. This eIF5 domain can bind simultaneously to eIF3–NIP1 and eIF2β, consistent with a role in bridging eIF2–eIF3 interactions. Mutation of the AA-boxes in eIF5 leads to dissociation of the multi-eIF complex in cell extracts and diminishes the fraction of ribosomes engaged in translation in vivo. These results suggest strongly that the multifactor complex is an important intermediate in translation initiation in yeast.
Results
Amino-terminal segment of eIF3 subunit NIP1 (p93) can interact simultaneously with eIF1 and eIF5 in vitro
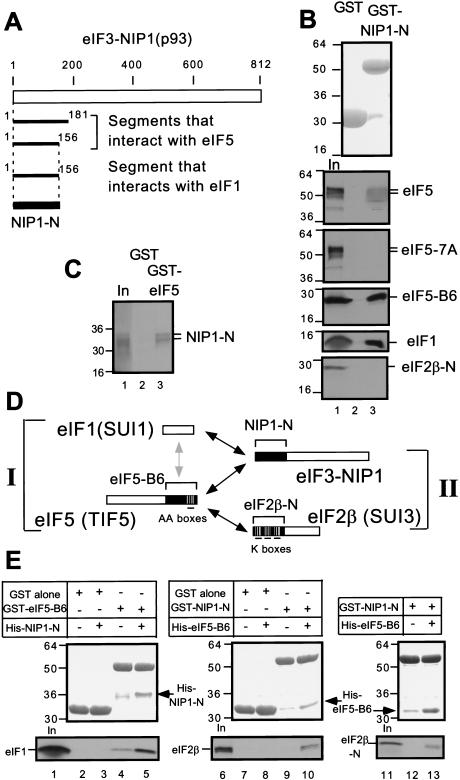
eIF1, eIF2, and eIF5 are implicated in correct recognition of AUG start codons in the yeast Saccharomyces cerevisiae (Huang et al. 1997), but the physical linkages among these factors have not been fully elucidated. Because the eIF3 subunit NIP1 bound both to eIF1 and eIF5 in vitro (Asano et al. 1998; Phan et al. 1998), we attempted to identify the segment of NIP1 responsible for these interactions. Toward this end, we screened a yeast two-hybrid library containing overlapping 300–600 nucleotide fragments derived from the genes encoding eIF3 subunits (Asano et al. 1998) for interactions with constructs encoding full-length eIF1 or eIF5. As shown in Figure 1A, we identified two different amino-terminal segments of NIP1 containing residues 1–181 and 1–156 using eIF5 as bait. We also obtained the smaller of these NIP1 segments (designated NIP1-N) using eIF1 as bait. We confirmed these results by in vitro binding assays using GST-fusions to either eIF5 (Asano et al. 1999) or NIP1-N expressed in E. coli and 35S-labeled NIP1-N, eIF5, or eIF1 synthesized in vitro. The 35S-NIP1-N fragment bound to GST–eIF5, but not to GST alone (Fig. 1C) and 35S-eIF1 and 35S-eIF5 both bound to GST–NIP1-N but not to GST alone (Fig. 1B).
Figure 1.
Simultaneous interactions in vitro involving minimal binding segments of eIF3–NIP1, eIF2β, eIF5, and full-length eIF1. (A) Summary of yeast two-hybrid interactions detected between segments of NIP1 and full-length eIF5 and eIF1 (see Materials and Methods for details). The empty box indicates the 812-amino acid NIP1 polypeptide. Segments of NIP1 that interacted with eIF5 and eIF1 are listed below with locations in the sequence. (B) and (C) The amino-terminal segment of eIF3–NIP1 (NIP1-N) binds to eIF1 and eIF5. 20μg of GST–NIP1-N (B, lane 3), GST–eIF5 (C, lane 3), or GST alone (B and C, lanes 2), expressed in Escherichia coli and immobilized on glutathione sepharose beads, was incubated with the 35S-labeled proteins listed on the right. After extensive washing, the bound proteins were analyzed by SDS-PAGE followed by Coomassie staining (B, top panel) and autoradiography (B, lower five panels; C). Lanes 1 of B and C, 50% of the input amounts of labeled proteins. (D) Schematic illustration of interactions involving full-length eIF1 and segments of eIF2, eIF3, and eIF5 indicated with brackets and designated as in the text. Boxes indicate primary structures of the proteins involved and the filled regions denote the minimal binding domains. Black or gray arrows indicate direct interactions; the gray arrow denotes a relatively weak interaction. (E) Bridging experiments. 5 μg of GST–eIF5-B6, GST–NIP1-N, or GST alone immobilized on beads was mixed with 35S eIF1 (lanes 2–5), 35S eIF2β (lanes 7–10), or 35S eIF2β-N (lanes 12,13) in the presence or absence (as indicated) of 60 μg of His–NIP1-N or His–eIF5-B6 expressed in E. coli and purified by Ni2+ affinity chromatography. Bound proteins were detected by SDS-PAGE, followed by Coomassie staining (middle) and autoradiography (bottom). The GST-fusion or His-tagged proteins used in each reaction are listed above each of the middle panels. In lanes 1, 6, and 11, 50% of the input amounts of 35S-proteins were loaded.
We also found that GST–NIP1-N bound specifically to the carboxy-terminal ∼40% of eIF5 (eIF5-B6), and failed to bind to mutant eIF5 bearing seven Ala substitutions in AA-box 2 (eIF5-7A) (Fig. 1B). GST–NIP1-N did not interact with the amino-terminal half of eIF2β (eIF2β-N), previously shown to bind to GST–eIF5 (Asano et al. 1999). These results indicate that the carboxy-terminal segment of eIF5 containing the AA-boxes is responsible for its binding to the amino-terminal 156 residues of NIP1.
Having found that the NIP1-N segment can interact with both eIF1 and eIF5, we wished to determine whether these interactions could occur simultaneously. For this purpose, we purified a polyhistidine-tagged form of the eIF5-B6 segment expressed in E. coli (His–eIF5-B6) and tested it for the ability to compete with 35S-eIF1 for binding to GST–NIP1-N. Addition of a 12-fold molar excess of His–eIF5-B6 to GST–NIP1-N did not reduce the amount of 35S-eIF1 bound to GST–NIP1-N (data not shown), suggesting that a stable ternary complex containing all three proteins, GST–NIP1-N/His-eIF5-B6/35S-eIF1, could be formed. To test this idea directly, we purified a polyhistidine-tagged form of NIP1-N (His–NIP1-N) and asked whether it could bridge an interaction between GST–eIF5-B6 and 35S-eIF1. As shown in Figure 1E, lane 4, 35S-eIF1 interacted weakly with GST–eIF5-B6 in the absence of His–NIP1-N. Addition of a 12-fold molar excess of His–NIP1-N to GST–eIF5-B6 substantially increased the amount of 35S-eIF1 bound to GST–eIF5-B6 (Fig. 1E, lane 5, bottom panel). As expected, a fraction of the His–NIP1-N fragment was recovered in the complexes containing GST–eIF5-B6 (Fig. 1E, lane 5, middle panel). We conclude that the NIP1-N segment can enhance interaction between eIF1 and eIF5 by its ability to bind simultaneously to each of these factors (summarized in Fig. 1D, I).
Carboxy-terminal one-third of eIF5 can interact simultaneously with eIF2β and eIF3–NIP1 in vitro
We showed previously that the carboxy-terminal segment of eIF5 (eIF5-B6) could interact separately with eIF3–NIP1 and eIF2β (Asano et al. 1999). The latter interaction was mapped to the amino-terminal half of eIF2β (eIF2β-N) and requires at least one of the three polylysine stretches (K boxes 1–3) present in that segment (Das et al. 1997; Asano et al. 1999) (summarized in Fig. 1D, II). Accordingly, we wished to determine whether eIF5-B6 could interact simultaneously with recombinant forms of NIP1-N and eIF2β-N. Addition of excess His–NIP1-N did not reduce the amount of complex formed between GST–eIF5-B6 and 35S-labeled eIF2β-N (data not shown), consistent with the idea that a His–NIP1-N/GST–eIF5-B6/35S-eIF2β-N ternary complex could be formed. To pursue this possibility further, we asked whether His–eIF5-B6 could bridge interactions between GST–NIP1-N and 35S-eIF2β or 35S-eIF2β-N. As shown in Figure 1E (lanes 9,10,12,13), 35S-eIF2β or 35S-eIF2β-N bound to GST–NIP1-N only when His–eIF5-B6 was present in the reaction. As expected, a fraction of the His–eIF5-B6 was recovered in the complexes containing GST–NIP1-N. These findings indicate that the carboxyl terminus of eIF5 can promote complex formation by NIP1 and eIF2β by interacting simultaneously with the amino-terminal domains of each factor (Fig. 1D, II).
The carboxyl terminus of eIF5 mediates association of eIF3 with eIF2 in vivo
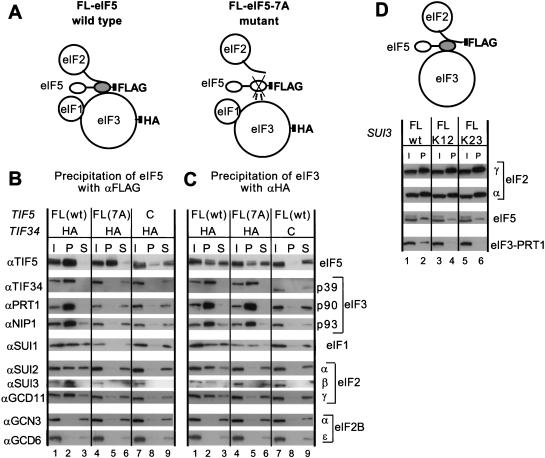
The results summarized in Figure 1D suggest that simultaneous binding of eIF3–NIP1 to eIF1 and eIF5, and the binding of eIF5 to both NIP1 and eIF2β, could lead to the formation of a multifactor complex containing eIF2, eIF3, eIF5, and eIF1. To investigate whether this occurs in vivo, we carried out immunoprecipitation experiments using extracts from yeast strains expressing FLAG-tagged eIF5 (FL-eIF5), either wild-type or bearing the tif5-7A mutation, and an HA-tagged form of the TIF34 subunit of eIF3 (HA-TIF34) (see Fig. 2A for experimental design). When the extracts were immunoprecipitated with anti-FLAG antibodies, the results we obtained confirmed our previous finding (Asano et al. 1999) that a large fraction of the eIF3 subunits (∼70%–100 %) and a smaller proportion of eIF2 subunits (∼10%–20%) were coimmunoprecipitated with eIF5 (Fig. 2B, lane 2). Furthermore, both eIF5–eIF3 and eIF5–eIF2 interactions were impaired by the tif5-7A mutation (Fig. 2B, lane 5), indicating their mutual dependence on AA-box 2. We also found that ∼20% of the eIF1 was coimmunoprecipitated specifically with FL-eIF5 in a manner dependent on AA-box 2. As expected, we did not observe coimmunoprecipitation with FL-eIF5 of the α or ε subunits of eIF2B, the guanine nucleotide exchange factor for eIF2 (Fig. 2B, lanes 2,5).
Figure 2.
Coimmunoprecipitation of eIF2 and eIF3 from cell extracts dependent on AA-box 2 in the carboxyl terminus of eIF5. (A) Schematic model of the structures of the multifactor complexes found in KAY50 and KAY51 (Table 1) containing wild-type eIF5-FL (left) or eIF5-FL-7A (right), as deduced from this study. The modular structure of eIF5 is represented by two ovals (amino-terminal and carboxy-terminal domains) connected with a thick line (less conserved region). The shaded oval in eIF5 corresponds to the B6 fragment, necessary and sufficient for binding to eIF2 and eIF3 (Asano et al. 1999). This oval is crossed in the mutant, indicating the 7A mutation in AA-box 2. The curved line attached to eIF2 represents the amino-terminal half of eIF2β containing the K-boxes which interact with the carboxy-terminal domain of eIF5. Filled squares indicate the epitope tags on eIF3-HA-TIF34 or FL-eIF5. Dotted lines indicate a weakened interaction. (B, C) Coimmunoprecipitation of eIFs with eIF5-FL or TIF34-HA. WCEs were prepared from strains KAY50, KAY51, KAY10, and KAY37 (Table 1), grown in YPD medium at 30°C. Aliquots of WCEs were incubated with anti-FLAG (B) or anti-HA (C) affinity resin and after extensive washing, the bound proteins were analyzed by SDS-PAGE and immunoblotting using the antibodies indicated on the left. Lanes 1, 4, and 7, 20% input (I) amounts of WCE; lanes 2, 5, and 8, the entire precipitated (P) fractions; lanes 3, 6, and 9, 10% of supernatant (S) fractions. (D) Coimmunoprecipitation of eIFs with eIF2β-FL. The schematic model is similar to that described in B and C except that eIF2β carries the FLAG epitope. WCEs prepared from strains KAY33 (SUI3), KAY25 (SUI3-FL), KAY29 (sui3-FL-K12) and KAY30 (sui3-FL-K23) (Asano et al. 1999) were immunoprecipitated with FLAG affinity resin and the precipitated proteins were analyzed by immunoblotting using the appropriate antibodies. (I) 20% input amount of WCE (lanes 1, 3, and 5); (P) the entire precipitated fractions (lanes 2, 4, and 6). K12 and K23 indicate the Ala substitutions present in K-boxes 1–2 or 2–3, respectively, in the mutant eIF2β-FL proteins.
The results just described do not distinguish between the presence of separate eIF5–eIF2 and eIF5–eIF3 complexes versus a multifactor complex containing all three factors. To make this distinction, the same extracts were treated with anti-HA antibodies to immunoprecipitate eIF3 and the immune complexes were probed for eIF2 subunits and eIF5. We found that 10%–20% of each eIF2 subunit was coimmunoprecipitated with HA-TIF34 from the extract bearing wild-type FL-eIF5 but not from that containing the FL-eIF5-7A mutant (Fig. 2C, lanes 2,5). These results indicate that a substantial proportion of eIF2 was associated with eIF3 in a manner dependent in vivo on AA-box 2 of eIF5.
As expected, eIF5 (∼20%) coimmunoprecipitated with HA-TIF34 (Fig. 2C); however, in contrast to the results in Figure 2B, this interaction was only reduced rather than abolished by the tif5-7A mutation. To explain this apparent discrepancy, we suggest that the tif5-7A mutation has a stronger effect on the interaction of eIF5 with eIF2 compared to eIF5–eIF3 association, as indicated by the results in Figure 2C. In the experiment shown in Figure 2B, the carboxyl terminus of eIF5 would be tethered to the FLAG antibody and this may have perturbed the conformation of the AA-box region in a manner that exacerbates the negative effect of the tif5-7A mutation on eIF5–eIF3 interaction, thus leading to complete dissociation of eIF3 from eIF5-7A (Fig. 2B). The coimmunoprecipitation of eIF1 with HA-TIF34 was ostensibly reduced by the tif5-7A mutation; however, the amount of this protein in the mutant extract was diminished to an even greater degree. Accordingly, the proportion of total eIF1 associated with eIF3 was not decreased by the AA-box 2 mutation in eIF5. This last finding is consistent with our previous conclusion that eIF1 interacts with eIF3 directly via eIF3–NIP1 (Asano et al. 1998) (Fig. 1). We found that expression of eIF1 was reduced consistently by the tif5-7A mutation, perhaps indicating that eIF1 is less stable when the physical association between eIF5 and other initiation factors is disrupted. This reduction in the amount of eIF1 is not responsible for the slow growth phenotype associated with tif5-7A (Asano et al. 1999), as overexpression of eIF1 did not suppress this phenotype (data not shown).
eIF5 binds to the amino-terminal domain of the β-subunit of eIF2 dependent on the three K-boxes in that region of the protein (Das et al. 1997; Asano et al. 1999). If eIF5 bridges an interaction between eIF2 and eIF3, as concluded above, we reasoned that mutations in the K-boxes that weaken eIF2–eIF5 association should simultaneously decrease eIF2–eIF3 interaction in vivo. To test this prediction, we immunoprecipitated FLAG-tagged eIF2β from extracts of strains expressing the tagged wild-type protein or tagged mutant proteins containing Ala substitutions in the Lys residues of K-boxes 1 and 2, or 2 and 3. As expected, a fraction of eIF5 and the PRT1 subunit of eIF3 were coimmunoprecipitated with wild-type FL-eIF2β (Fig. 2D, lane 2). Mutating K-boxes 1–2 or 2–3 in FL-eIF2β reduced coimmunoprecipitation of eIF5 by a factor of ∼3, as reported previously (Asano et al. 1999), and abolished coimmunoprecipitation of eIF3–PRT1 with FL-eIF2β (Fig. 2D, lanes 2,4,6). Thus, we conclude that substantial fractions of the eIF2, eIF3, eIF5, and eIF1 in cell extracts reside in the same high molecular weight complexes whose integrity depends on AA-box 2 in eIF5 and the K-boxes in eIF2β.
eIF1, eIF2, eIF3, and eIF5 form a multifactor complex free of 40S ribosomes
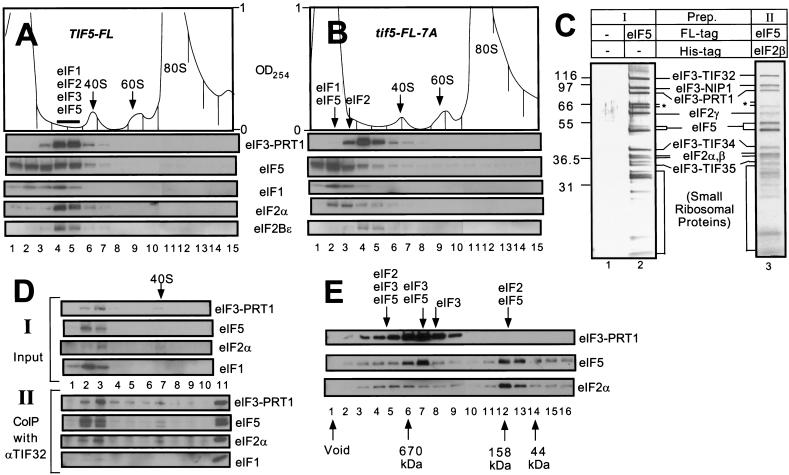
It was possible that the multifactor complex postulated above was stable only when associated with the 40S ribosome. In an effort to detect the complex free of ribosomes, whole cell extracts (WCEs) from yeast strains expressing FL-eIF5 or FL-eIF5-7A were resolved by sucrose gradient-velocity sedimentation and the fractions were analyzed by immunoblotting. As shown in Figure 3A for the FL-eIF5 extract, the majority of eIF2α and eIF3–PRT1, and roughly one-half of the eIF1 cosedimented in two fractions (fractions 4–5) behind the 40S ribosome peak (fractions 6–7). A large proportion of the FL-eIF5 was additionally present in fractions 4–5, with the remainder sedimenting more slowly in fractions 1–3. The eIF2Bε analyzed as a control was present in fractions 4–5, presumably representing the eIF2B–eIF2 holocomplex (Cigan et al. 1993; Asano et al. 1999). In the extract containing FL-eIF5-7A, only a small fraction of the eIF5, eIF1, and eIF2 sedimented in fractions 4–5, and the majority of these factors was found in fractions 1–3 (Fig. 3B). In addition, the position of eIF3–PRT1 was shifted slightly towards the top of the gradient. In contrast, the sedimentation rate of eIF2Bε (and a small proportion of the eIF2α) was unaffected by the tif5-7A mutation (Fig. 3B).
Figure 3.
The multifactor complex containing eIF1, eIF2, eIF3, and eIF5 can be isolated free of 40S ribosomes. (A,B) Twenty A260 units of WCEs prepared from KAY50 (TIF5-FL) (A) and KAY51 (tif5-FL-7A) (B) were resolved on 15%–40% sucrose gradients by centrifugation for 4.5 h at 39,000 rpm. Gradients were separated into 20 0.6 mL-fractions while scanning continuously at A254 as shown ( top panels). Portions of the fractions were precipitated with 10% TCA and analyzed by SDS-PAGE and immunoblotting using antibodies against the factors listed beside the five bottom panels. (C) Silver staining of purified preparations of the multifactor complex. The staining patterns of Type I (lane 2) and Type II (lane 3) preparations are shown along with a mock preparation obtained from a strain containing untagged eIF5 (lane 1) in parallel with the Type I preparation. (*) Degradation products of TIF32. (D) Size fractionation and coimmunoprecipitation analysis of a Type II preparation of the multifactor complex. A sample containing 0.5 mL (50 μg protein) of Type II preparation was resolved on 7.5%–30% sucrose gradient by centrifugation for 5 h at 41,000 rpm. The gradient was separated into 10 1.2 mL-fractions while scanning continuously at A254. One hundred μL of each fraction were precipitated with 10% TCA (top panels, input), 400 μL were immunoprecipitated with anti-TIF32 antibodies (bottom panels, CoIP with αTIF32), and the resulting samples were analyzed by immunoblotting using antibodies against the factors listed to the right. The position of the 40S ribosomes is indicated. (Lane 11) 1% of the Type II preparation separated on the sucrose gradient. (E) Size fractionation of Type I preparation of the multifactor complex. Two hundred microliters of Type I preparation was separated on a Superose 6 sizing column (Pharmacia), preequilibrated with buffer A without pepstatin, aprotinin, and leupeptin, using the FPLC system (Pharmacia), as described previously (Phan et al. 1998). Portions of the fractions were precipitated with 10 % TCA, and analyzed by SDS-PAGE and immunoblotting using antibodies against the factors listed on the right. Lane 1: sample from the void volume. Arrows below the panels indicate the elution positions of size standards (BioRad) determined in a parallel experiment. Arrows above the panels indicate the positions of different complexes deduced from the immunoblot analysis.
The results in Figure 3B strongly support the notion that a multifactor complex containing eIF1, eIF2, eIF3, and eIF5 can exist free of the 40S ribosome and is dependent on the bipartite motif in eIF5 for its integrity. In the FL-eIF5-7A extract, it appeared that this complex dissociated into free eIF2 (124 kD; fractions 2–3), eIF3 (∼600 kD; fractions 3–5), eIF5 (50 kD; fractions 1–3), and eIF1 (13 kD; fractions 1–3) (Fig. 3B). Given that eIF1 interacts weakly with eIF5 (Fig. 1E), it is possible that eIF1 dissociated from eIF3 during centrifugation due to the impaired interaction between eIF3 and FL-eIF5-7A. We suspect that all of the residual eIF2 detected in fractions 4–5 for the FL-eIF5-7A extract is associated with eIF2B (Fig. 3B).
We attempted to purify the multifactor complex by affinity chromatography followed by size fractionation. Two different strategies for affinity purification of the complex were employed. One preparation (Type I) was obtained by one-step affinity purification directed against FL-eIF5 using FLAG antibody resin, and was conducted with yeast extracts containing wild-type eIF3–TIF34 (strain KAY37) or the HA-tagged form of this protein (strain KAY50). The second preparation (Type II) was obtained by two-step affinity purification directed firstly against a polyhistidine-tagged form of eIF2β using Ni2+ affinity resin, and secondly against FL-eIF5 using FLAG antibody resin, using extracts from strain KAY56 (TIF5-FL SUI3-His). Silver-staining (Fig. 3C, lanes 2,3) and Western blot analysis (data not shown) of both affinity-purified preparations prior to size fractionation revealed that eIF5 and the subunits of eIF2 and eIF3 accounted for the majority of the higher molecular mass constituents, whereas no eIF2B subunits were detected. The Type I preparation contained substoichiometric amounts of eIF4G1, but the more highly purified Type II complex did not (data not shown). Substantial amounts of lower molecular mass proteins (<32 kD) also were present. The banding pattern of the latter closely resembled that of isolated 40S ribosomal subunit proteins (data not shown), suggesting that a sizable proportion of the FL-eIF5 and other eIFs had copurified with 40S ribosomal subunits. As expected, none of these proteins was present in a Type I preparation obtained from an isogenic strain harboring untagged eIF5 (Fig. 3C, lane 1).
We were able to separate the eIFs from the 40S ribosomal subunits in the Type II preparation by sucrose gradient-velocity sedimentation. Western blot analyses (Fig. 3D, top panel) and silver staining (data not shown) of the gradient fractions revealed that most of the eIF1, eIF2, eIF3, and eIF5 sedimented in fractions 2–3, whereas the 40S ribosomes were present in fractions 6–7. To determine whether the multifactor complex was retained after separation from the 40S ribosomes, we immunoprecipitated the gradient fractions with monoclonal antibodies against eIF3 subunit TIF32 and probed the immune complexes for other factors. As shown in the bottom four panels of Figure 3D, almost all of the eIF5, eIF2α, and eIF3–PRT1 in fractions 2 and 3 were coimmunoprecipitated with eIF3–TIF32. Physical linkage of eIF2 and eIF3 is the hallmark of the multifactor complexes described above containing these two factors bridged by the carboxy-terminal domain of eIF5. Thus, we conclude that high molecular mass complexes containing eIF2, eIF3, and eIF5 can persist in the absence of 40S ribosomes.
Only a small fraction of eIF1 in fractions 2–3 was immunoprecipitated by the TIF32 antibodies. As eIF1 binds to eIF3–NIP1, which in turn interacts with eIF3–TIF32 (Asano et al. 1998), the TIF32 antibodies may have perturbed the eIF1 binding domain in eIF3, leading to its dissociation from the multifactor complex during the immunoprecipitation. Additionally, the association of eIF1 with eIF3 is inherently more labile than that of the eIF3–eIF5 interaction (Phan et al. 1998).
Gel filtration of the affinity-purified Type I preparation on a Superose 6 column confirmed the presence of a high molecular mass complex containing eIF3, eIF5, and eIF2; however, a mixture of partial complexes were additionally observed in this experiment (Fig. 3E). Most of the eIF2 was present in a complex with an apparent Mr of ∼158,000 that contained eIF5 but was devoid of eIF3 (Fig. 3E, fractions 12,13). A similar eIF2–eIF5 complex has been observed in mammalian cell extracts (Chaudhuri et al. 1994). A second peak of eIF2 present in fractions 4–6 had a much higher apparent Mr (∼1.1 × 106) and also contained eIF5 and eIF3. We suggest that these latter fractions contain the multifactor complexes containing eIF2, eIF3, and eIF5 detected by the coimmunoprecipitation experiments described above. As 40S ribosomes elute from this column in the void volume (data not shown), these findings provide additional evidence that the multifactor complex can persist independently of the ribosome.
In the experiment shown in Figure 3D, a large proportion of eIF2 was coimmunoprecipitated from the sucrose gradient fractions with eIF3 subunit TIF32. Given that a much smaller proportion of eIF2 coeluted with eIF3 from the Superose 6 column shown in Figure 3E, we presume that a substantial fraction of the eIF2/eIF3/eIF5 complexes in the starting Type I preparation dissociated into free eIF3 (fractions 8,9) and eIF2/eIF5 complexes (fractions 12,13) during gel filtration, possibly as a result of dilution. The peak Superose 6 fraction containing the majority of eIF3 and eIF5 (fraction 7) had an apparent Mr of ∼6 × 105 and a relatively low content of eIF2. It most likely represents the eIF3/eIF5 binary complexes we detected previously as the major form of eIF3 in cell extracts (Phan et al. 1998). These binary complexes would be expected to occur in the Type I preparation because it was affinity purified using only FLAG antibody resin directed against FL-eIF5. Nearly all of the eIF1 was present in fractions 15 and 16 of the column (data not shown), indicating that eIF1 readily dissociated from the eIF2/eIF3/eIF5 complex even though it consistently copurified with FL-eIF5 through FLAG affinity chromatography (Figs. 2B and 3D,I).
Taken together, the results in Figure 3 provide strong biochemical evidence for the existence of a multifactor complex containing eIF1, eIF2, eIF3, and eIF5 that can occur independently of ribosomes. Compared to the multisubunit factors eIF2 and eIF3, this multifactor complex appears to be labile, however, and its stability may be enhanced by independent interactions between the constituent eIFs and the 40S ribosome.
The multi-eIF complex is associated with initiator tRNAiMet in vivo
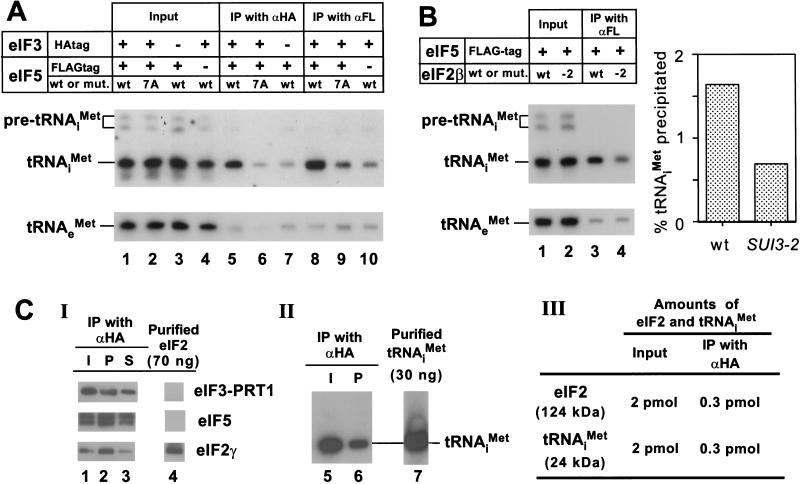
If the high molecular mass complexes described above containing eIF2 represent intermediates in the translation initiation pathway, then we would expect to find that initiator tRNAiMet could be coimmunoprecipitated from cell extracts with antibodies against eIF5 or eIF3, and that this interaction would be abolished by the tif5-7A mutation in AA-box 2 of eIF5. To test this prediction, we immunoprecipitated FL-eIF5 or HA-TIF34 from extracts of the strains described above expressing either wild-type or 7A mutant forms of FL-eIF5, and probed the immune complexes for initiator tRNA by Northern blot analyses. As shown in Figure 4A, tRNAiMet was coimmunoprecipitated specifically with FL-eIF5 or HA-TIF34 (eIF3) (Fig. 4A, lanes 5 and 8, respectively), and this interaction was reduced to background levels by the tif5-7A mutation (Fig. 4A, cf. lanes 6 and 9 with lanes 7 and 10). These findings suggest strongly that the multifactor complex contains tRNAiMet.
Figure 4.
The multifactor complex contains initiator tRNAiMet. (A) The eIF5-FL/eIF3-HA complex contains tRNAiMet. Samples containing 1 mg of WCE in 1 mL of buffer A (Asano et al. 1999) prepared in the presence of 0.2 U/μL of RNasin (Promega) from strains used in Figure 2B,C were incubated for 2 h at 4°C with 100 μL of Protein A Sepharose (Pharmacia) that was preincubated with 15 μL of mouse monoclonal anti-HA antibodies (BAbCO) or M2 FLAG affinity resin (Sigma). After washing with 1 mL buffer A four times, the beads were suspended in water, extracted twice with phenol and once with phenol/chloroform, and precipitated with ethanol. Precipitated RNAs were separated by denaturing polyacrylamide gel electrophoresis, followed by Northern blot analysis using probes specific for tRNAiMet or tRNAeMet, as described previously (Anderson et al. 1998) (lanes 5–10). Lanes 1–4, RNA samples phenol-extracted and ethanol-precipitated from 2% of the input WCEs used for the immunoprecipitations. The top panel describes the antibodies used for immunoprecipitation, the presence of epitope tags on eIF5 or eIF3–TIF34, and the presence of wild-type (wt) or tif5-7A (7A) forms of eIF5 in the extracts. (B) Evidence that tRNAiMet is bound to eIF2 in the multifactor complex. One milligram aliquots of WCEs prepared from strains KAY56 (SUI3) and KAY57 (SUI3-2) were incubated at 37°C for 5 min and subjected to immunoprecipitation with FLAG affinity resin, followed by Northern blotting, as described above (lanes 3 and 4, respectively). Lanes 1 and 2, RNA samples phenol-extracted and ethanol-precipitated from 2% of the input WCEs used for the immunoprecipitations. The histogram on the right shows the percentages of the amounts of tRNAiMet present in the starting extracts that were immunoprecipitated based on phosphorimaging analysis of the Northern data. (C) Quantification of the amounts of eIF2 and tRNAiMet in the multifactor complex. A 200 μL aliquot (20 μg) of Type I preparation was immunoprecipitated for 2 h at 4°C with 20 μL Protein A Sepharose beads that had been preincubated with 3 μL of anti-HA antibodies. (Panel I) After washing the beads four times with buffer A, the immunoprecipitated proteins were subjected to immunoblot analysis as described in Figure 2C, along with 70 ng of purified eIF2 (lane 4). Lane 1, 20 μL starting Type I preparation; lane 3, 10 % of supernatant fraction from the immunoprecipitation. (Panel II) RNA was extracted from a duplicate immunoprecipitation conducted as in Panel I and subjected to Northern blot analysis as described above (A), along with 30 ng purified tRNAiMet (lane 7). Lane 5, RNA extracted from 100 μL of the starting Type I preparation. (Panel III) The results of quantification of data in panels I–II.
It was important to demonstrate that the tRNAiMet associated with the multi-eIF complex is bound to eIF2, rather than to one of the other initiation factors in the complex that contains RNA-binding activity, such as eIF3–TIF35 (Hanachi et al. 1999). Towards this end, we asked whether the association between tRNAiMet and eIF5 would be disrupted by a mutation in eIF2β (SUI3-2) that was shown to reduce Met-tRNAiMet-binding to eIF2 in vitro (Huang et al. 1997). We immunoprecipitated FL-eIF5 from strains expressing wild-type (SUI3) or mutant (SUI3-2) forms of eIF2β and probed the immune complexes for tRNAiMet. Because the SUI3-2 mutant is temperature-sensitive for growth, we incubated both extracts at 37°C for 5 min prior to immunoprecipitation. As shown in Figure 4B, we observed ∼60% less tRNAiMet associated with eIF5-FL in the SUI3-2 versus the SUI3 extract. We also confirmed that interaction between eIF2 and eIF5-FL was not affected by the SUI3-2 mutation (data not shown). These results support the idea that the tRNAiMet associated with the multifactor complex is bound to eIF2.
To determine the stoichiometry of eIF2 and tRNAiMet in the multifactor complex, we compared the amounts of these components that were immunoprecipitated with anti-HA antibodies (and thus associated with HA-TIF34 in eIF3) with defined amounts of purified eIF2 and tRNAiMet. The results in Figure 4C indicate that similar amounts of eIF2 and tRNAiMet were present in the Type I preparation purified by FLAG affinity chromatography directed against FL-eIF5. Additionally, similar proportions of eIF2 and tRNAiMet coimmunoprecipitated with HA-TIF34. Based on these results, we conclude that nearly all the eIF2 physically associated with eIF3 in the multifactor complex is associated with tRNAiMet. As only the GTP-bound form of eIF2 binds Met-tRNAiMet (Trachsel 1996), this finding implies that the multifactor complex contains the eIF2/GTP/Met-tRNAiMet ternary complex.
Disruption of the multifactor complex diminishes translation initiation in vivo
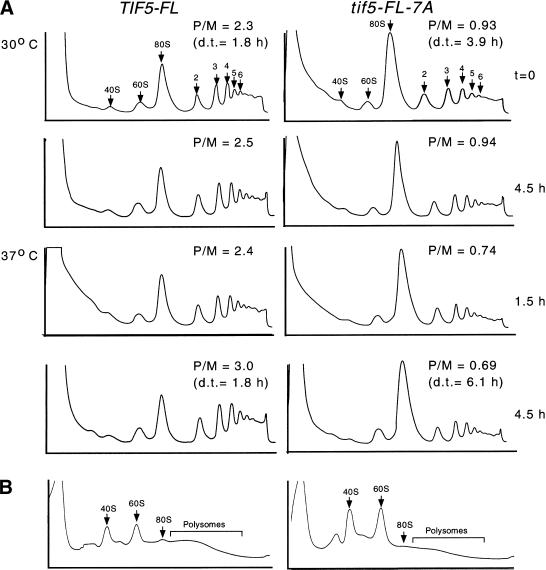
The tif5-7A mutation in AA-box 2 of eIF5 impairs the integrity of the multifactor complex in cell extracts. Hence, to assess the importance of the multifactor complex for efficient translation initiation in vivo, we examined the polysome profiles in cell extracts prepared from isogenic tif5-FL-7A and TIF5-FL strains. The mutant cells are temperature sensitive for growth and have a doubling-time in liquid medium that is approximately twofold and threefold greater than that of the wild-type at 30°C and 37°C, respectively (data not shown). Before harvesting the cells and preparing extracts, cycloheximide was added to immobilize translating 80S ribosomes on the mRNAs. As expected, the wild-type cells at 30°C had a sizable polysome content, characterized by a polysome to monosome ratio (P/M) >2.0, which increased to a value of 3.0 after 4.5 h at 37°C (Fig. 5A, left panels). In contrast, the P/M ratio for the tif5-7A mutant at 30°C was only 0.93 and was diminished further to ∼0.7 after 1.5 h or 4.5 h of incubation at 37°C (Fig. 5A, right panels).
Figure 5.
Analysis of polysome profiles in isogenic TIF5 and tif5-7A strains. (A) Yeast strains KAY50 and KAY51 grown exponentially in YPD medium at 30°C (t = 0) were shifted to 37°C (or kept at 30°C as a control) for the indicated times. Cycloheximide was added to the cultures for 5 min prior to harvesting the cells. WCEs were prepared and resolved by velocity-sedimentation on 5%–45% sucrose gradients. Fractions were collected while scanning continuously at A254. The positions of 40S and 60S subunits, 80S ribosomes and polysomes are indicated. (P/M) Ratio of A254 in the combined polysome fractions to that in the 80S peak; (d.t.) cell doubling time in hours. (B) WCEs prepared from yeast strains grown exactly as described for top panels in A were centrifuged on 5%–45% sucrose gradients containing 0.7 M NaCl for 2.5 h at 39,000 rpm and analyzed as in A.
To determine whether the ribosomal subunits accumulated in the tif5-7A mutant as translating 80S monosomes or as vacant 80S couples lacking mRNA, we repeated the experiment just described using a high salt buffer during centrifugation, in which 80S couples dissociate into free 40S and 60S subunits (Foiani et al. 1991). As shown in Figure 5B, the tif5-7A mutant had substantially elevated levels of free 40S and 60S subunits, even at 30°C (Note that the different polysomal species are not resolved under these buffer conditions). These findings indicate a substantial reduction in the frequency of translation initiation in the tif5-7A mutant, resulting in a large pool of vacant 80S ribosomes. Accordingly, we propose that the carboxy-terminal domain of eIF5 stimulates translation initiation in a manner dependent on the conserved AA boxes by promoting the stable association of initiation factors present in the multifactor complex.
Discussion
Previously, we had shown that eIF5 was associated with eIF3 and eIF2 in cell extracts, but it was unclear whether these proteins were bound to eIF5 in the same complexes. The carboxy-terminal domain of recombinant eIF5 had interacted separately with purified eIF2 and recombinant eIF2β, and also with purified eIF3 and recombinant eIF3–NIP1. As both interactions with eIF5 were dependent on the AA-boxes at the carboxyl terminus of eIF5, it was questionable whether these interactions could occur simultaneously (Asano et al. 1999). Here we presented biochemical evidence that the carboxyl terminus of eIF5 can bridge interactions between eIF2β and eIF3–NIP1 in vitro. Additionally, we found that the amino terminus of NIP1 can interact simultaneously with eIF1 and the carboxyl terminus of eIF5 (Fig. 1E). The occurrence of these simultaneous interactions between initiation factors led to the prediction of a multifactor complex containing eIF1, eIF3, eIF5, and eIF2.
We provided strong evidence for the presence of this complex in vivo by showing that eIF2 could be coimmunoprecipitated with eIF3 from cell extracts, dependent on AA-box 2 in the carboxyl terminus of eIF5 (Fig. 2). We also showed by velocity sedimentation analysis and biochemical purification techniques that the multifactor complex can exist free of 40S ribosomes (Fig. 3), and we discovered that it contains nearly stoichiometric amounts of eIF2 and tRNAiMet (Fig. 4). This last finding implies that the eIF2 present in the complex occurs in the ternary complex with GTP and Met-tRNAiMet.
The tif5-7A mutation in AA-box 2 at the carboxyl terminus of eIF5 destabilized the multifactor complex in cell extracts (Figs. 2C and 3B) and, importantly, led to a decrease in polysome content and an accumulation of vacant 80S couples in vivo (Fig. 5). Thus, it appears that the integrity of the multifactor complex is important for high level translation initiation in vivo. This idea is consistent with our previous finding that overexpression of all three subunits of eIF2 together with tRNAiMet restored eIF5–eIF2 physical association and partially suppressed the Ts− phenotype of a tif5-7A mutant (Asano et al. 1999). The tif5-7A mutation strongly inhibits growth at elevated temperatures but is not lethal (Asano et al. 1999). It appears that this mutation does not completely abolish the multifactor complex in vivo, as immunoprecipitation with antibodies against HA-tagged eIF3–TIF34 revealed residual eIF3–eIF5 interaction in the tif5-FL-7A extract (Fig. 2C). It is possible that a more disruptive mutation in the eIF5 carboxyl terminus that completely eliminates formation of the multifactor complex would be lethal.
The ability of eIF1, eIF3, eIF5, and the ternary complex to associate independently of the 40S ribosome raises the possibility that these factors bind to the 40S ribosome as a preformed unit. This could facilitate rapid assembly of 43S preinitiation complexes containing these factors, requiring only the addition of eIF1A and the mRNA/eIF4F/PAB1 complex to produce a 48S complex capable of locating the AUG start codon and hydrolyzing the GTP bound to eIF2. Mammalian eIF3 can bind to 40S ribosomes in the absence of other factors, although it has been reported that binding of eIF3 and eIF2 is synergistic (Benne and Hershey 1978; Peterson et al. 1979; Trachsel and Staehelin 1979; Chaudhuri et al. 1997). There is ample evidence from yeast and mammalian systems that eIF3 enhances binding of the ternary complex to 40S ribosomes; however, no physical contact between eIF2 and eIF3 has been detected. By bridging interaction between eIF3 and the ternary complex, the carboxyl terminus of eIF5 might promote the ability of eIF3 to stabilize ternary complex binding to the 40S ribosome. This hypothesis is not incompatible with the possibility that eIF3 stimulates ternary complex binding indirectly by allosteric alteration of the eIF2 binding site on the ribosome, or by preventing 60S subunit joining and formation of 80S couples.
Several observations from mammalian cell-free translation systems are ostensibly at odds with the idea that eIF5 promotes ternary complex binding to the 40S subunit. In partial initiation reactions reconstituted with purified factors, eIF5 displayed no ability to stimulate Met-tRNAiMet ternary complex binding to the 40S subunit (Trachsel et al. 1977; Benne and Hershey 1978). However, it is likely that the eIF5 used in those experiments was the factor now recognized as eIF5B, which promotes 60S subunit joining following hydrolysis of GTP by eIF2 (Pestova et al. 2000) and the eIF5 may have been introduced with eIF3. Secondly, the purified mammalian eIFs—eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, and eIF4F—are sufficient for assembly of a 48S complex on mRNA with the Met-tRNAiMet positioned at the AUG codon (Pestova et al. 1998). It is conceivable that the hypothetical contribution of eIF5 to 48S complex formation is redundant with the activities of other factors, in which case formation of the multifactor complex identified here could be rate-enhancing rather than essential for production of 48S complexes in vivo.
In current models of the initiation pathway, eIF5 is shown interacting transiently with the 48S preinitiation complex to stimulate GTP hydrolysis by eIF2 only after Met-tRNAiMet base pairs with the AUG start codon (Merrick and Hershey 1996). It could be argued that the most serious defect in tif5-7A mutants is an impairment of eIF5 GAP function due to diminished interaction of soluble monomeric eIF5 with eIF2 bound to the 48S preinitiation complex. This defect would explain the Ts− phenotype conferred by tif5-7A independently of its deleterious effect on eIF5–eIF3 interaction and the integrity of the multifactor complex. Consistent with this possibility, multiple substitutions in acidic residues of the AA boxes in rat eIF5 that impaired binding to recombinant eIF2β diminished eIF5 GAP activity in vitro and reduced the growth rate of yeast cells expressing the rat proteins in place of endogenous eIF5. However, certain mutations nearly abolished interaction between rat eIF5 and eIF2β but only produced a modest reduction in GAP activity (Das and Maitra 2000). Our preliminary experiments have revealed no effect of the tif5-7A mutation on yeast eIF5 GAP activity in vitro (data not shown). Moreover, our previous finding that overexpressing the ternary complex suppressed partially the tif5-7A mutation (Asano et al. 1999) is difficult to explain if impaired binding of monomeric eIF5 to 40S-bound ternary complexes is solely responsible for the Ts− phenotype of tif5-7A mutants. Producing more ternary complexes in the cell should not promote binding of monomeric eIF5 to 48S preinitiation complexes that already contain the ternary complex. Indeed, the presence of excess ternary complexes could interfere with GTP hydrolysis in preformed 48S complexes by titrating eIF5 into nonribosomal complexes. Hence, we favor the idea that overexpressing the ternary complex stimulated translation initiation in tif5-7A cells by restoring formation of the multifactor complex, enabling eIF5 to enter the preinitiation complex early in the pathway in association with other factors involved in start site selection.
The presence of eIF5 and eIF1 in a multifactor complex containing eIF3 and the ternary complex is consistent with genetic and biochemical studies in yeast implicating eIF1, eIF5, and all three subunits of eIF2 in stringent AUG selection. Our finding that eIF5 and eIF1 can bind simultaneously to the amino-terminal 20% of eIF3–NIP1 suggests that these factors are tethered in proximity on the surface of eIF3. The fact that eIF5 can interact with eIF2 concurrently with eIF3–NIP1 further suggests that all of the factors involved in stringent AUG selection are juxtaposed in the 48S initiation complex by the multiple protein–protein interactions detected in our studies. Loss of this precise spatial organization among factors on the ribosome resulting from the tif5-7A mutation might substantially reduce the rate of GTP hydrolysis following AUG recognition by Met-tRNAiMet, leading to dissociation of 48S complexes and the failure to join with the 60S subunit.
Determining the relative contributions of the eIF5 carboxyl terminus and its multiple interactions with other eIFs will require extensive characterization of the effects of tif5-7A and other mutations in the eIF5 carboxyl terminus on the different steps of the initiation pathway, both in vitro and in vivo. The fact that the amino acid sequences of the eIF5 carboxyl terminus and its interacting domains in eIF2β and eIF3–NIP1 are conserved between yeast and mammals (Asano et al. 1997, 1999) indicates that appreciating the full significance of these interactions and their role in stabilizing the multifactor complex is an important goal for future research.
Materials and methods
Plasmid construction, yeast strains, and two-hybrid analysis
pGAD–NIP1-N, encoding the GAL4 activation domain fused to the amino-terminal 156 amino acids of eIF3–NIP1, was obtained multiple times by two-hybrid screening of a library of pGAD424 fusions to randomly selected fragments of ∼100–200 amino acids from the five subunits of eIF3, as described previously (Asano et al. 1998). The GAL4 DNA binding fusions to eIF1 or eIF5 encoded by pGBT–SUI1 (Asano et al. 1998) and pGBT–TIF5 (Asano et al. 1999), respectively, were employed as bait. pGEX–NIP1-N and pHis–NIP1-N were constructed by transferring the NIP1 insert of pGAD–NIP1-N to pGEX–4T-1 (Smith and Johnson 1988) and pET15b (Novagen), respectively, for production of the corresponding GST and polyhistidine-tagged fusions to the amino-terminal 156 amino acids of NIP1. DNA segments encoding full-length wild-type eIF5 or eIF5-7A (Asano et al. 1999) and the amino-terminally truncated eIF5 derivative known as B6 (Asano et al. 1999) were inserted behind the T7 promoter in pT7-7 (Tabor and Richardson 1987) or pET23a (Novagen) to generate pT7–TIF5, pT7–TIF5-7A, and pET–TIF5-B6, respectively, and were used to synthesize the corresponding 35S-labeled polypeptides in vitro. pHis–TIF5-B6 encoding polyhistidine-tagged eIF5-B6 under the T7 promoter was constructed by inserting the TIF5-B6 DNA fragment behind the T7 promoter in pET15b. Other plasmids used in this study for the production of various forms of yeast eIFs were described previously (Asano et al. 1998, 1999; Phan et al. 1998). S. cerevisiae strains produced for this study are listed in Table 1, and details of their construction are available upon request.
Table 1.
Yeast strains employed in this study
| Strain
|
Genotype
|
|---|---|
| KAY50 | MATa leu2-3, -112 ura3-53 trp1-Δ63 gcn2Δ tif34Δ::hisG tif5Δ::hisG p[TIF5-FL TRP1] p[TIF34-HA LEU2] |
| KAY51 | MATa leu2-3, -112 ura3-53 trp1-Δ63 gcn2Δ tif34Δ::hisG tif5Δ::hisG p[tif5-FL-7A TRP1] p[TIF34-HA LEU2] |
| KAY37 | MATa leu2-3, -112 ura3-53 trp1-Δ63 gcn2Δ tif5Δ::hisG p[TIF5-FL TRP1] |
| KAY10 | MATa leu2-3, -112 ura3-53 trp1-Δ63 gcn2Δ tif34Δ::hisG p[TIF34-HA LEU2] |
| KAY56 | MATa leu2-3, -112 ura3-53 trp1-Δ63 sui3Δ tif5Δ::hisG p[TIF5-FL TRP1] p[SUI3-His LEU2] |
| KAY57 | MATa leu2-3, -112 ura3-53 trp1-Δ63 sui3Δ tif5Δ::hisG p[TIF5-FL TRP1] p[SUI3-His-2 LEU2] |
Protein purification
Polyhistidine-tagged forms of NIP1-N and eIF5-B6 were expressed in E. coli BL21(DE3) from the pHis–NIP-N and pHis–TIF5-B6 plasmids, respectively, and purified with Ni2+ affinity resin (Promega) as recommended by the manufacturer. eIF2 and tRNAiMet were purified as described previously (Pavitt et al. 1998; Anderson et al. 2000).
The multifactor complex was affinity-purified as follows. For Type I preparations, ∼10 mg of WCEs from strain KAY37 (TIF5-FL) or KAY50 (TIF5-FL TIF34-HA) in buffer A (Asano et al. 1999) was mixed with 100 μL of M2 FLAG-affinity resin (Sigma) for 2 h at 4°C. The bound proteins were eluted in 200 μL of buffer A containing 400 ng/μL FLAG peptide (Sigma), after washing the beads with 1 mL buffer A four times. The resulting FL-eIF5 preparation contained ∼100 ng/μL the multi-factor complex. For Type II preparations, ∼1 g of WCE, prepared from strain KAY56 (TIF5-FL SUI3-His) in buffer Imd10 (20 mM Tris-HCl at pH 7.5, 50 mM KCl, 10 mM MgCl2, 10% glycerol, 10 mM imidazole, 5 mM NaF, 7 mM β-mercaptoethanol, 1 mM PMSF, Complete protease inhibitors [Boehringer Manheim], 1 μg/mL each of pepstatin, leupeptin, and aprotinin) was mixed with 5 mL Ni2+-agarose (Qiagen) for 1 h at 4°C. After washing the beads twice with 30 mL buffer Imd20 (same as Imd10 except containing 20 mM imidazole), crude His–eIF2β-containing complex was eluted in 10 mL Imd 200 (same as Imd10 except containing 200 mM imidazole). This preparation, plus 30 mL buffer A as above, was then mixed with 250 μL M2 FLAG-affinity resin for 2 h at 4°C. The bound proteins were eluted in 500 μL of buffer A containing FLAG peptide, as described above for Type I preparations. The resulting His–eIF2β/FL-eIF5 Type II preparation contained ∼100 ng/μL of the multifactor complex. Purification to this extent was necessary for efficient coimmunoprecipitation of eIF2 with anti-TIF32 eIF3 antibodies (Valasek et al. 1998) after separation of the multifactor complex in sucrose gradient-velocity sedimentation, as shown in Figure 3D.
Biochemical assays
Immunoprecipitations of the multifactor complex from WCEs with antibodies against HA- or FLAG (FL)-epitopes were conducted in buffer A as described (Asano et al. 1998, 1999). Protein–protein interactions between recombinant forms of yeast eIFs were analyzed as described (Asano et al. 1999).
WCEs or the Type II multifactor preparation was resolved on sucrose gradients, prepared in buffer K (20 mM Tris-HCl at pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM NaF, 1 mM DTT, 1 mM PMSF, Complete protease inhibitors, 1 μg/mL each of pepstatin, leupeptin, and aprotinin), in 14 × 89 mm polyallomer tubes (Beckman) by centrifugation with a Beckman SW41 rotor. Gradient samples were fractionated with an ISCO gradient fraction collector. For the coimmunoprecipitations in Figure 3D, 400 μL of each fraction were mixed with 10 μL of GammaBind Plus Sepharose (Pharmacia) that was preincubated with 2 μL of mouse monoclonal anti-TIF32 antibodies (Valasek et al. 1998), and after washing the beads with buffer A four times, the bound proteins were analyzed by immunoblotting. These TIF32 antibodies specifically immunoprecipitated eIF1, eIF3, and eIF5 from yeast WCEs (L. Valasek and A.G. Hinnebusch, unpubl.).
For polysome analysis, 300 mL of yeast cells grown to OD600 = 0.5 ∼ 2 and treated with 50 μg/mL cycloheximide for 5 min were harvested by centrifugation, washed in 5 mL ice-cold buffer K containing 50 μg/mL cycloheximide and 200 μg/mL heparin, and resuspended in a 1.5-fold packed cell volume of buffer K containing the same concentrations of cycloheximide and heparin. WCEs were prepared by homogenizing the washed cells by vortexing with glass beads as described previously (Garcia-Barrio et al. 1995). Fifteen A260 units of WCEs were layered on 5%–45% sucrose gradient prepared in a buffer (20 mM Tris-HCl at pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM DTT) and centrifuged for 2.5 h at 39,000 rpm in a Beckman SW41 rotor. The positions of ribosomal species were determined by scanning the gradient at A254 with the ISCO gradient fraction collector.
Acknowledgments
We are indebted to Tom Donahue for his timely and kind gifts of materials used in this study. We also credit Jim Anderson with the initial observation of association between eIF3–HA-TIF34 and tRNAiMet and thank him for providing purified yeast tRNAiMet. We also thank Thanuja Krishnamoorthy and Leos Valasek for providing purified eIF2 and monoclonal anti-TIF32 antibodies, respectively, and Minerva Garcia-Barrio for advice on polysome analysis. We thank Tom Dever and Lon Phan for critical reading of the manuscript and members of the Hinnebusch and Dever laboratories for helpful discussion and technical advice. A.S. was supported by an Endocrine Fellowship from the Diabetes Branch, NIDDK, NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ahinnebusch@nih.gov; FAX (301) 496-6828.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad. 831800.
References
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes & Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Phan L, Hinnebusch AG. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci. 2000;97:5173–5178. doi: 10.1073/pnas.090102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Kinzy TZ, Merrick WC, Hershey JWB. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Chem. 1997;272:1101–1109. doi: 10.1074/jbc.272.2.1101. [DOI] [PubMed] [Google Scholar]
- Asano K, Phan L, Anderson J, Hinnebusch AG. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:18573–18585. doi: 10.1074/jbc.273.29.18573. [DOI] [PubMed] [Google Scholar]
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP–GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Maitra U. Cloning and characterization of the p42 subunit of mammalian translation initiation factor 3 (eIF3): Demonstration that eIF3 interacts with eIF5 in mammalian cells. Nucl Acids Res. 1999;27:1331–1337. doi: 10.1093/nar/27.5.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, Hershey JWB. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- Castilho-Valavicius B, Yoon H, Donahue TF. Genetic characterization of the Saccharomyces cerevisiae translational initiation suppressors sui1, sui2 and SUI3 and their effects on HIS4 expression. Genetics. 1990;124:483–495. doi: 10.1093/genetics/124.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Das K, Maitra U. Purification and characterization of bacterially expressed mammalian translation initiation factor 5 (eIF-5): Demonstration that eIF-5 forms a specific complex with eIF-2. Biochemistry. 1994;33:4794–4799. doi: 10.1021/bi00182a007. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Chakrabarti A, Maitra U. Biochemical characterization of mammalian translation initiation factor 3 (eIF3) J Biol Chem. 1997;272:30975–30983. doi: 10.1074/jbc.272.49.30975. [DOI] [PubMed] [Google Scholar]
- Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE. Promotion of Met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- Cigan AM, Pabich EK, Feng L, Donahue TF. Yeast translation initiation suppressor sui2 encodes the α subunit of eukaryotic initiation factor 2 and shares identity with the human α subunit. Proc Natl Acad Sci. 1989;86:2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan AM, Bushman JL, Boal TR, Hinnebusch AG. A protein complex of translational regulators of GCN4 is the guanine nucleotide exchange factor for eIF-2 in yeast. Proc Natl Acad Sci. 1993;90:5350–5354. doi: 10.1073/pnas.90.11.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaie P, Wittmer B, Altmann M, Trachsel H. Isolation of a protein complex containing translation initiation factor Prt1 from Saccharomyces cerevisiae. J Biol Chem. 1995;270:4288–4292. doi: 10.1074/jbc.270.9.4288. [DOI] [PubMed] [Google Scholar]
- Das S, Maitra U. Mutational analysis of mammalian translation initiation factor 5 (eIF5): Role of interaction between the β-subunit of eIF2 and eIF5 in eIF5 function in vitro and in vivo. Mol Cell Biol. 2000;20:3942–3950. doi: 10.1128/mcb.20.11.3942-3950.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Maiti T, Das K, Maitra U. Specific interaction of eukaryotic translation initiation factor 5 (eIF5) with the β-subunit of eIF2. J Biol Chem. 1997;272:31712–31718. doi: 10.1074/jbc.272.50.31712. [DOI] [PubMed] [Google Scholar]
- Donahue TF, Cigan AM, Pabich EK, Castilho-Valavicius B. Mutations at a Zn(II) finger motif in the yeast elF-2β gene alter ribosomal start-site selection during the scanning process. Cell. 1988;54:621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- Dorris DR, Erickson FL, Hannig EM. Mutations in GCD11, the structural gene for eIF-2γ in yeast, alter translational regulation of GCN4 and the selection of the start site for protein synthesis. EMBO J. 1995;14:2239–2249. doi: 10.1002/j.1460-2075.1995.tb07218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg B, McLaughlin CS, Moldave K. Analysis of temperature-sensitive mutant ts187 of Saccharomyces cerevisiae altered in a component required for the initiation of protein synthesis. J Biol Chem. 1982;257:10846–10851. [PubMed] [Google Scholar]
- Fletcher CM, Pestova TV, Hellen CUT, Wagner G. Structure and interactions of the translation initiation factor eIF1. EMBO J. 1999;18:2631–2639. doi: 10.1093/emboj/18.9.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barrio MT, Naranda T, Cuesta R, Hinnebusch AG, Hershey JWB, Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes & Dev. 1995;9:1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- Hanachi P, Hershey JWB, Vornlocher HP. Characterization of the p33 subunit of eukaryotic translation initiation factor-3 from Saccharomyces cerevisiae. J Biol Chem. 1999;274:8546–8553. doi: 10.1074/jbc.274.13.8546. [DOI] [PubMed] [Google Scholar]
- Huang H, Yoon H, Hannig EM, Donahue TF. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes & Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC, Hershey JWB. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey JWB, Matthews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- Naranda T, MacMillan SE, Hershey JWB. Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. J Biol Chem. 1994;269:32286–32292. [PubMed] [Google Scholar]
- Naranda T, MacMillan SE, Donahue TF, Hershey JW. SUI1/p16 is required for the activity of eukaryotic translation initiation factor 3 in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2307–2313. doi: 10.1128/mcb.16.5.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Ramaiah KVA, Kimball SR, Hinnebusch AG. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes & Dev. 1998;12:514–526. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Peterson DT, Merrick WC, Safer B. Binding and release of radiolabeled eukaroytic initiation factors 2 and 3 during 80 S initiation complex formation. J Biol Chem. 1979;254:2509–2519. [PubMed] [Google Scholar]
- Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Tabor S, Richardson CC. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel H. Binding of initiator methionyl-tRNA to ribosomes. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 113–138. [Google Scholar]
- Trachsel H, Staehelin T. Initiation of mammalian protein synthesis: The multiple functions of the initiation factor eIF-3. Biochim Biophys Acta. 1979;565:305–314. doi: 10.1016/0005-2787(79)90207-7. [DOI] [PubMed] [Google Scholar]
- Trachsel H, Erni B, Schreier MH, Staehelin T. Initiation of mammalian protein synthesis: The assembly of the initiation complex with purified initiation factors. J Mol Biol. 1977;116:755–767. doi: 10.1016/0022-2836(77)90269-8. [DOI] [PubMed] [Google Scholar]
- Valasek L, Trachsel H, Hasek J, Ruis H. Rpg1, the Saccharomyces cerevisiae homologue of the largest subunit of mammalian translation initiation factor 3, is required for translational activity. J Biol Chem. 1998;273:21253–21260. doi: 10.1074/jbc.273.33.21253. [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Donahue TF. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]