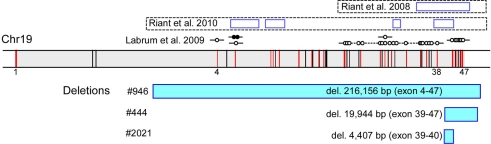
Figure 1.
Newly identified and previously reported EA2-causing genomic rearrangements in CACNA1A that vary in size and location. In a diagram of the genomic structure of CACNA1A on chr 19 spanning approximately 300, 000 base pairs (shaded in gray), the vertical bars represent exons, and those in red harbor target sequences for the MLPA probes. The locations of exons 1, 4, 38, and 47 are specified. The rectangles highlighted in blue below the genomic structure of CACNA1A represent missing MLPA probes in the patients, labeled with the sizes and deleted exons. The open rectangles above the genomic structure of CACNA1A represent previously described deletions, with established boundaries, in five unrelated kindreds with EA2 (Riant et al., 2008, 2010), along with additional instances of genomic rearrangements without breakpoint ascertainment by D:Labrum et al., 2009; open circle – missing MLPA probe; closed circle – duplicated MLPA probe.