Figure 3.
Phasing Inheritance-State Blocks by Parsimony
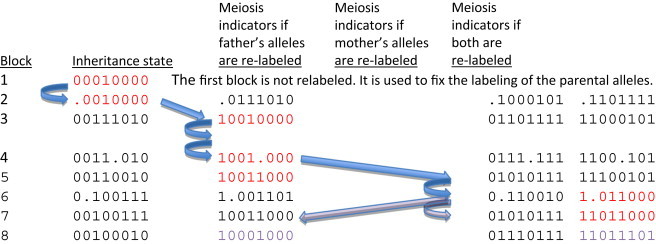
An inheritance-state vector for four children of a sextet consists of 8 bits. The first, third, fifth, and seventh bits relate the paternal alleles of each of the four children, and the second, fourth, sixth, and last bit relate the maternal alleles. If two bits are identical (i.e., 0 and 0 or 1 and 1), the alleles are IBD. If the bits are not identical (e.g., 1 and 0), the alleles are not IBD. If one of the bits is the ambiguity character (•) then IBD is not determined between that pair of individuals. By convention, the first two bits of an inheritance-state vector are always set to 0. Inheritance-state vectors can be converted to meiosis-indicator vectors by relabeling the bits for each block so that they consistently correspond to the meiotic origin of each allele, rather than simply relating IBD status between individuals. There are four possible meiosis-indicator vectors for each inheritance-state vector. Adjacent blocks of the genome are separated by short distances between informative variants that localize recombinations and so the parsimonious choice of the four labelings is the one that minimizes the number of recombinations between adjacent states. If there has been a single recombination, there is exactly one choice of labeling that represents a single recombination from the previous block (blue arrows). If there are two or more recombinations, then there could be more than one parsimonious choice and ambiguity results (purple arrows). The set of meiosis-indicator vectors in red corresponds to the parsimonious labelings that reflect one recombination each between blocks 1 and 3, 3 and 5, and 5 and 7. Blocks 2, 4, and 6 are intervals in which recombinations have occurred and so contain an ambiguity character.
