Figure 1.
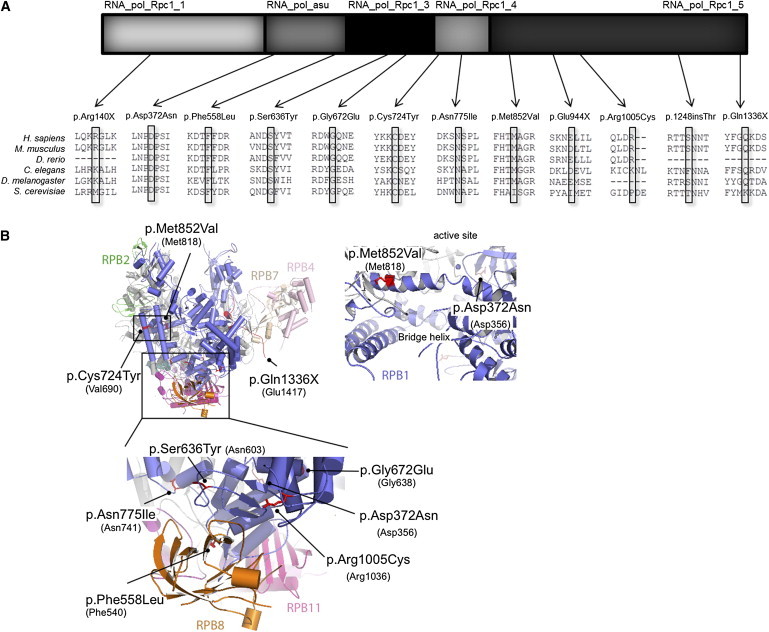
POLR3A Mutations
(A) Schematic representation of POLR3A exonic mutations. Missense and nonsense mutations are depicted according to the different POLR3A (RPC1) domains (modified from Ensembl). The amino acid conservation across species is shown for all missense and nonsense mutations. The following abbreviations are used in the upper panel: RNA_pol_Rpc1_1, RNA_pol_Rpc1_3, RNA_pol_Rpc1_4, and RNA_pol_Rpc1_5, RNA polymerase subunit domains one, three, four and five; and RNA_pol_asu, RNA polymerase alpha subunit. The following abbreviations are used in the lower panel: C. elegans, Caenorhabditis elegans; D. melanogaster, Drosophila melanogaster; D. rerio, Danio rerio; H. sapiens, Homo sapiens; M. musculus, Mus musculus; and S. cerevisiae, Saccharomyces cerevisiae.
(B)Three-dimensional representations of POLR3A missense and nonsense mutations. Point mutations identified in POLR3A are displayed according to their equivalent positions in the yeast RNA Polymerase II RPB1 subunit.27 Numbers in brackets refer to the positions in yeast RPB1. Images have been generated with PyMol software (Schrödinger). For imaging purposes, the RPB5 subunit has been omitted from the figure. RPB1 is shown in blue, RPB2 in green, RPB7 in brown, RPB4 in pink, RPB8 in orange, and RPB11 in magenta.
