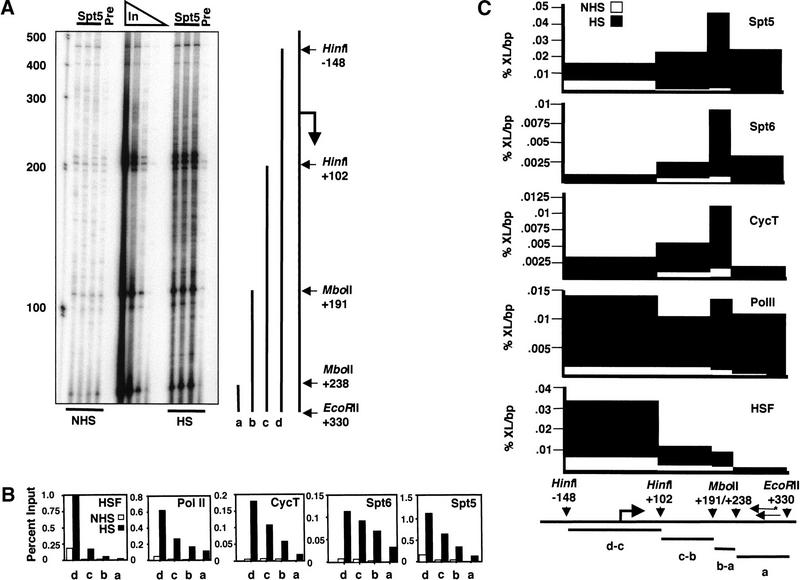
Figure 6.
High-resolution determination of protein occupancy on hsp70. (A) Ligation-mediated PCR analysis on hsp70. hsp70 restriction endonuclease sites and fragments created by partial digestion at HinfI, and MboII sites are shown to right of the gel, and these define one end of the region analyzed: EcoRII was used to cut the DNA to completion and thereby define the other end of the region analyzed. The fragments produced by this digest are shown: a, b, c, and d. The complete analysis, starting with the immunoprecipitation step, was performed in triplicate, and the different samples show very good agreement for all bands that were quantified. “Pre” is an immunoprecipitation performed with a preimmune serum control. (B) Determination of amount of hsp70 DNA immunoprecipitated relative to input DNA. Bands were quantified, and background signals from the preimmune controls were subtracted in each case. The percentage of the total sample that is immunoprecipitated for each of the four fragments is plotted. The triplet shown at HinfI +102 (see A) results from small-length polymorphisms among the hsp70 genes, and the signal from the entire triplet was used to quantify this fragment. Note that HSF and Pol II antibodies coimmunoprecipitate more DNA than do Spt6 and cyclin T antibodies (note the different scales for the abscissae). (C) Determination of protein density on hsp70. The densities were calculated from the measurements in part B. The densities from each interval were derived from subtraction of the next shortest fragment (except for a, which is the shortest), and then dividing by the length of the DNA interval. Note the different scales for the abscissae. (%XL) Percentage crosslinked immunoprecipitated DNA relative to input; (bp) base pair; (*) 32P end-label.