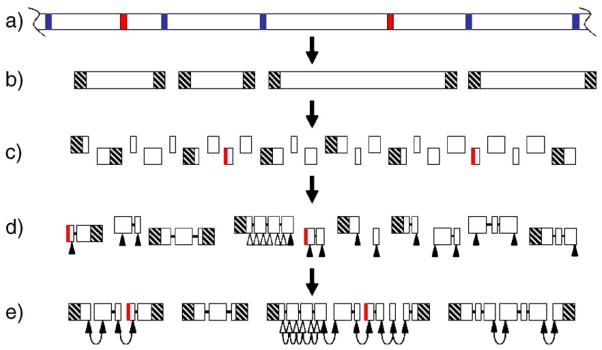
Fig. 1.
The genome of Tetrahymena thermophila and strategy for the assembly of the macronuclear genome. (a) The micronuclear genome consists of large chromosomes (only part of one chromosome is shown). Copies of these chromosomes are cut at CBSs (blue), and IESs (red) are spliced out, to produce the smaller macronuclear chromosomes (b), which are capped with telomeres (hatched). (c) Shotgun sequencing of the macronuclear chromosomes yields many sequence contigs, some of which may consist of—or be prematurely truncated by the incorporation of—IESs from contaminating micronuclear DNA (red vertical lines). (d) Contigs can be linked to form scaffolds using read-pair information (heavy black lines). Sequences close to nontelomeric scaffold ends (filled triangles) plus some from within larger scaffolds (open triangles) are chosen for use as markers. (e) HAPPY mapping reveals physical links between markers, allowing the scaffolds to be grouped into superscaffolds (hoops indicate HAPPY links) and also confirming the correct assembly within some larger scaffolds.