Figure 1.
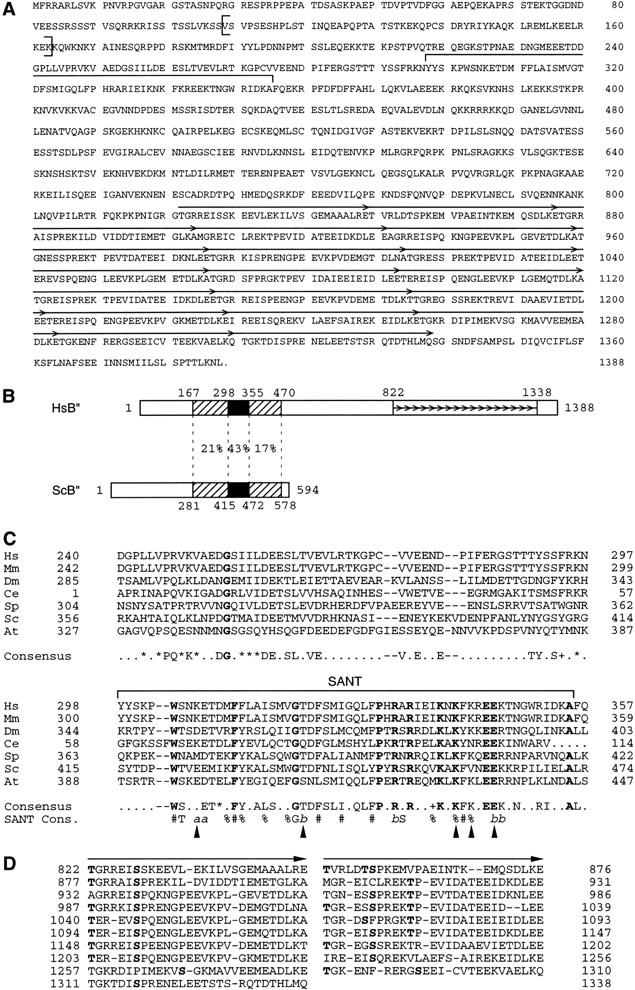
Structure of a hB′′. (A) Predicted hB′′ protein sequence (GenBank accession number AF298151). The SANT domain is indicated by a large horizontal bracket above the sequence. The repeated sequences are indicated by arrows. The sequence between the vertical brackets (amino acids 109–163) is lacking in some cDNAs that probably correspond to splicing variants. (B) Regions of similarity in the Homo sapiens and S. cerevisiae B′′ sequences (HsB′′ and ScB′′, respectively). The black boxes correspond to the SANT domain; the hatched boxes indicate regions of lower but still significant similarity on either side of the SANT domain. The percentages indicate identities between the two proteins in the regions delimited by the dotted lines. The numbers above the boxes indicate aa numbering. The small arrows indicate the repeats in hB′′. (C) Alignment of the SANT domain and region immediately upstream from hB′′ (Hs) and ScB′′ (Sc), as well as putative mouse (Mm), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Schizosaccharomyces pombe (Sp), and Arabidopsis thaliana (At) B′′ homologs. The SANT domain is indicated by the horizontal bracket above the sequence. The region N-terminal of the SANT domain was shown, in ScB′′, to be required for efficient transcription of linear yeast U6 templates in vitro (Kassavetis et al. 1998). The amino acids in boldface are identical in all sequences. The amino acids indicated in the consensus are identical in at least four of the seven sequences. (+) similar amino acids in all sequences; (*) similar amino acids in at least five of the seven sequences. The following amino acids were considered similar: V, I, L, and M; D and E; N and Q; R, K, and H; S and T; and W, F, and Y. The SANT consensus sequence (Aasland et al. 1996) is also shown. In the SANT consensus: (%) semiconserved hydrophobicity; (#) strongly conserved hydrophobicity; (a) conserved acidic residues; and (b) conserved basic residues. The positions marked with arrowheads indicate positions not conserved in the B′′ and SANT consensus sequences. The GenBank accession numbers are as follows: Mm ESTs, AI315861, AI315862, and AI787462; Ce genomic sequences U97016 (B0261) (nucleotides 25059–25185 and 25518–25780); Dm protein, AAF53291.1 (CG9305 gene product); Sp protein, T41239; and At protein T08564 (hypothetical protein T22F8.60). The numberings correspond to the sequence shown in (A) for HsB′′, to the sequences published (Kassavetis et al. 1995; Roberts et al. 1996; Rüth et al. 1996) for ScB′′, and to the sequences corresponding to the protein accession numbers indicated for other sequences. For the Mm sequence, the numbering corresponds to a putative mouse B′′ N-terminal region (amino acids 1–377) assembled from an alignment of various ESTs (GenBank accession numbers AI527964, AI317698, AI787358, AI526613, AI316225, AI639984, AA168265, AW456745, AW049390, AI429656, AI430369, AI450927, W91010, AI787462, AI315861, AI315862, AI892036, and AI315863), and for the Ce sequence, the numbering is arbitrary. (D) Alignment of the repeats within the C-terminal half of hB′′. As indicated by the arrows, each repeat can itself be subdivided into two short imperfect repeats. Residues in boldface indicate potential phosphorylation sites for various kinases, including PKC, PKA, CAMII, and CKII kinases.
