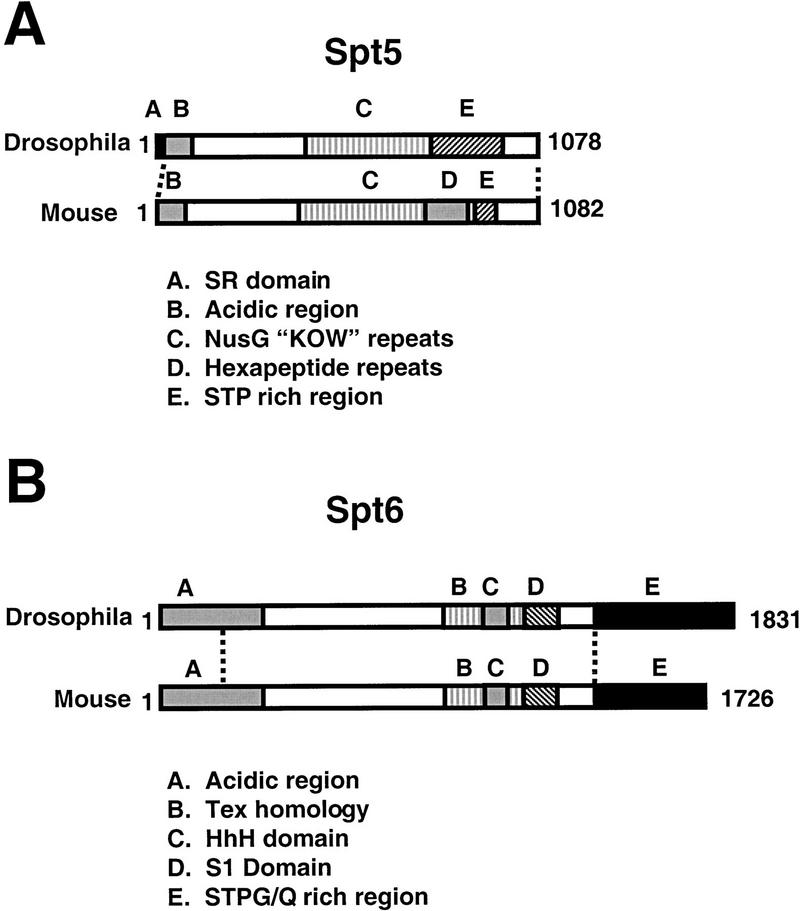
Figure 1.
Conservation and domain structure of the Drosophila melanogaster Spt5 and Spt6 proteins. Overall regions of homology indicated by dashed lines. (A) Drosophila Spt5 domain structure illustrated with murine Spt5 as a comparison. Spt5 proteins have acidic amino termini (region B), sequence homology to Escherichia coli NusG (region C) (Hartzog et al. 1998; Wada et al. 1998a; Wu-Baer et al. 1998), and serine-, threonine-, and proline-rich carboxy-terminal repeat regions noted in Yamaguchi et al. (1999b) and defined as CTR1 and CTR2 in Stachora et al. (1997) (regions D and E). Region E of Drosophila Spt5 has characteristics of CTR1 and CTR2 but the repeats appear degenerate. Homology of D. melanogaster Spt5 determined by BLAST (Altschul et al. 1990) with murine Spt5 is 50% amino-acid identity (E value = 0.0) and with Saccharomyces cerevisiae is 26% amino-acid identity (E value = 1e-58). The amino-terminal RS domain of D. melanogaster (region A) is novel for the Spt5 proteins. (B) D. melanogaster Spt6 domain structure illustrated with murine Spt6 as a comparison. Like Spt5 proteins, Spt6 proteins have acidic amino-termini (region A). Spt6 proteins also have sequence homology with a prokaryotic family of proteins implicated in transcription regulation (region B, named after the Bordetella pertussis Tex protein (Fuchs et al. 1996) and this region contains a conserved helix–hairpin–helix fold (HhH, region C, [Doherty et al. 1996]). Most Spt6 proteins are also predicted to contain RNA-binding S1 domains (region D, [Bycroft et al. 1997]). All Spt6 proteins except S. cerevisiae Spt6 have extended, divergent carboxyl termini rich in certain amino acids such as serine, threonine, and glycine (Drosophila) or glutamine (mouse) (region E and data not shown). Homology (by BLAST) of D. melanogaster Spt6 with murine Spt6 is 48% amino-acid identity (E value = 0.0) and S. cerevisiae Spt6 is 22% amino-acid identity (E value = 3e-79).