Figure 4.
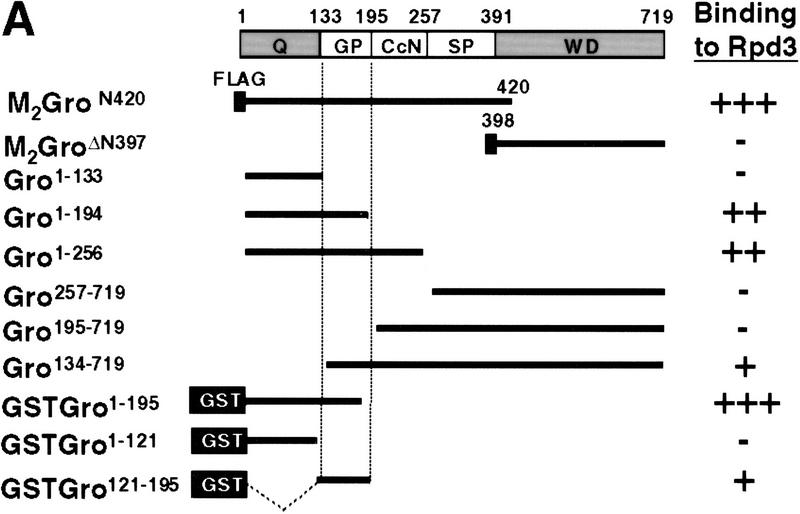
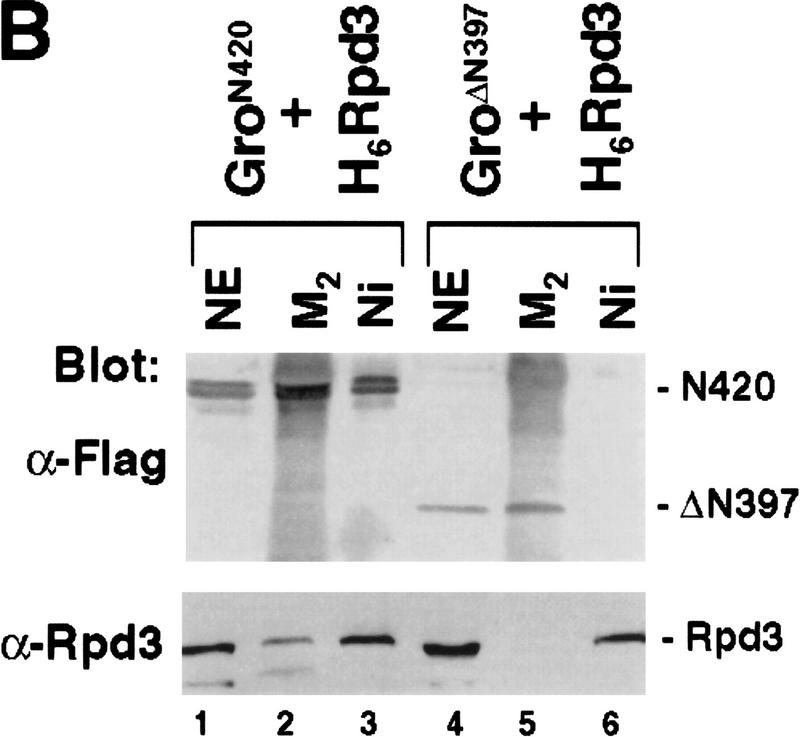
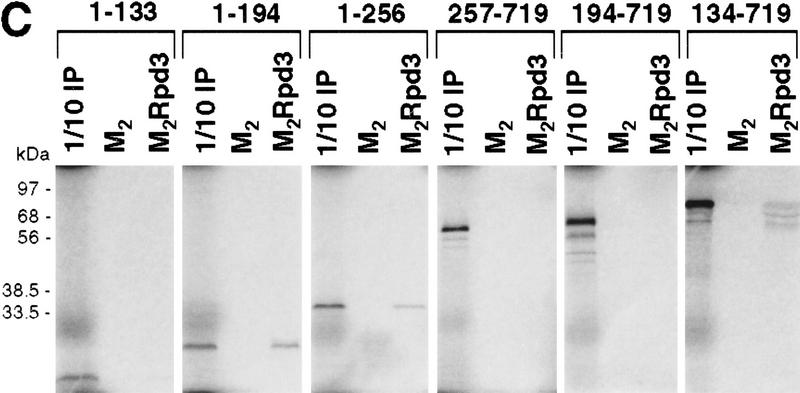
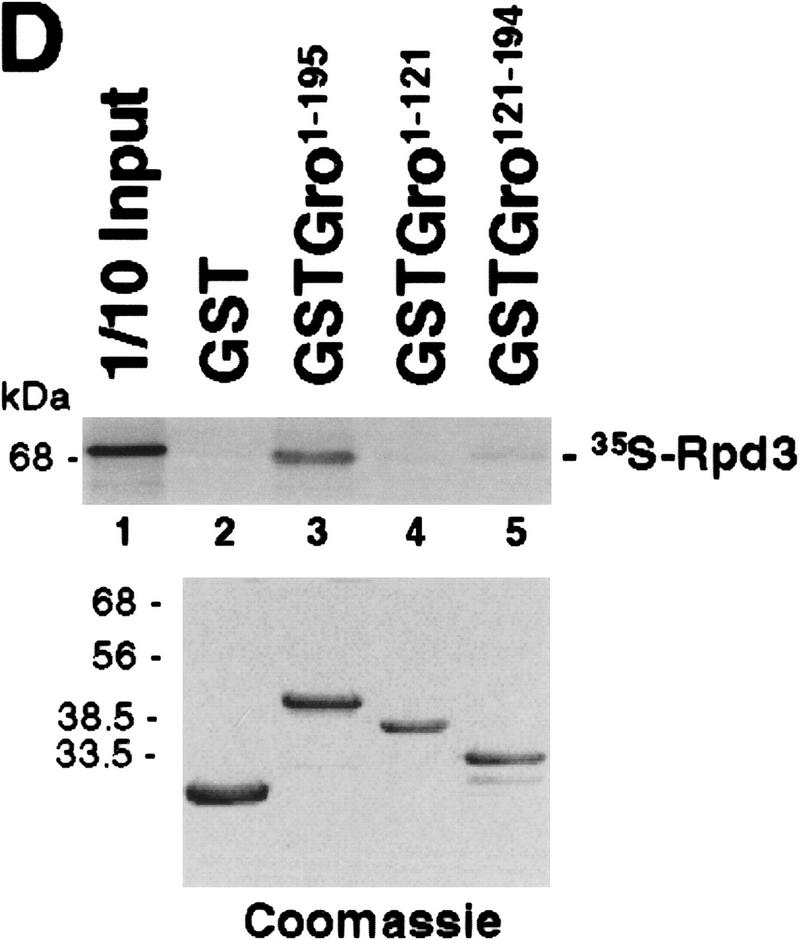
Mapping the Rpd3-interaction domain to the GP region of Gro. (A) Schematic diagram of different Gro deletions used in the protein interaction assays described in B–D. The conserved glutamine-rich (Q) and WD repeat (WD) domains of Gro are shaded. GP and SP denote the glycine/proline and serine/proline rich regions of Gro. CcN represents the motif containing putative cdc2 and casein kinase II phosphorylation sites as well as a nuclear localization signal. (B) The WD-repeat domain of Gro is dispensable for the Rpd3 interaction. Equal amounts of nuclear extracts prepared from insect cells coexpressing a Flag-tagged Gro deletion (M2GroN420 or M2GroΔ397) and H6Rpd3 were affinity purified using Ni2+–NTA–agarose (Ni) or anti-FLAG affinity beads (M2). Purified proteins were immunoblotted with anti-FLAG (top) or anti-Rpd3 (bottom) antibodies. (C) The GP domain of Gro is required for the interaction with Rpd3. Equal amounts of anti-Flag affinity beads alone (M2) or beads containing purified M2Rpd3 were used to examine the interac-tions with the indicated 35S-labeledGro deletions. Bound proteins were resolved by SDS-PAGE and visualized by autoradiography. (1/10 IP) Ten percent of each 35S-labeled protein input. (D) GST pull-down assays confirm the requirement of the GP domain for the Rpd3 interaction. Highly purified GST–Gro fusions were immobilized on glutathione beads and incubated with 35S-labeled Rpd3. After extensive washing of the beads, bound proteins were eluted and separated by SDS-PAGE and visualized by autoradiography (top). (Bottom) A Coomassie blue stained SDS–polyacrylamide gel of the purified GST–Gro fusions.