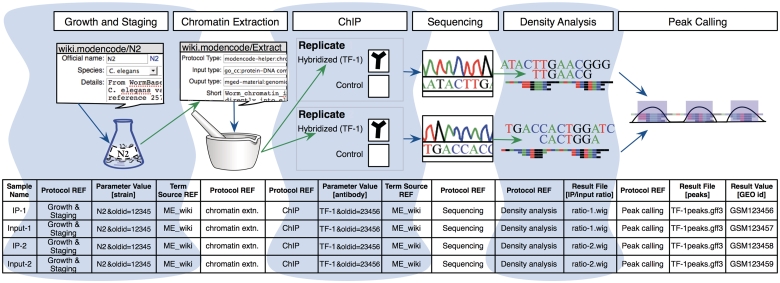
Figure 2.
A model experiment submitted to the modENCODE DCC and its mapping to metadata components BIR-TAB SDRF and the wiki. The top half is a diagram of experimental steps for a model ChIP-seq experiment: a worm culture is prepared, the genomic DNA associated with chromatin is extracted, followed by division of the extraction into two biological replicates. These are further subdivided, with half of each DNA sample used as a control, while the other is exposed to a specific TF antibody in a ChIP step. The resulting materials are prepared for sequencing, and the data processed to identify the set of binding sites occupied by the TF tested. The corresponding BIR-TAB SDRF is shown in the bottom half, and mirrors the flow of experimental steps as indicated by the green (output) and blue (input) arrows. The inputs and outputs are the arcs connecting each protocol node of an experiment represented in the database. Each cell in a protocol column of the BIR-TAB file maps to a specific wiki page where the inputs and outputs of that protocol have been indicated. Most experimental parameters, such as strain and antibody, are also specified in the wiki. A reference to the wiki for these experimental parameters or results is indicated with a Term Source REF column immediately following the parameter column.