Fig. 1.
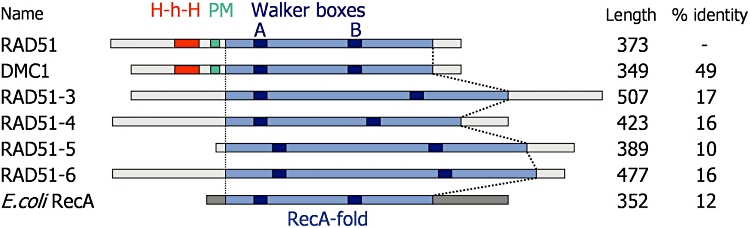
RAD51, DMC1 and RAD51 paralogues in T. brucei. Primary structures of T. brucei RAD51-related proteins relative to each other and to Escherichia coli RecA are shown; the proteins' lengths (amino acids) and sequence identity with T. brucei RAD51 are indicated. A conserved core (RecA-fold; light blue) is present in all proteins, containing Walker A and B motifs (dark blue). All the T. brucei proteins, except RAD51-5, contain N-terminal extensions that are not conserved with RecA; in this region RAD51 and DMC1 encode helix–hairpin–helix (H–h–H) and polymerization (PM) motifs not found in the RAD51 paralogues. A C-terminal extension in RecA is conserved with other bacteria RecA proteins, but is not conserved with the RAD51-related proteins.
