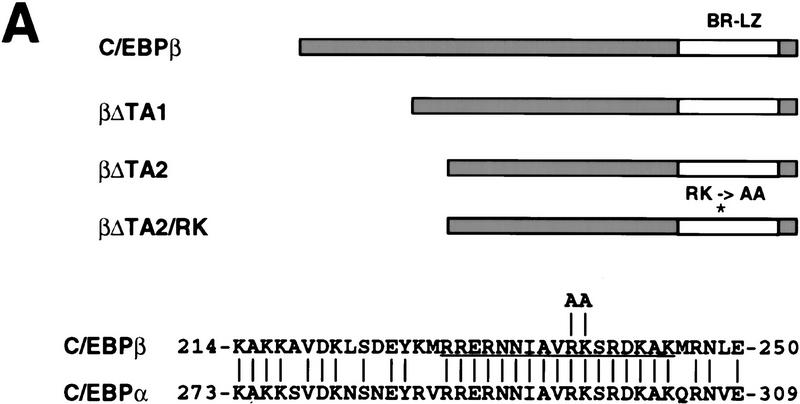
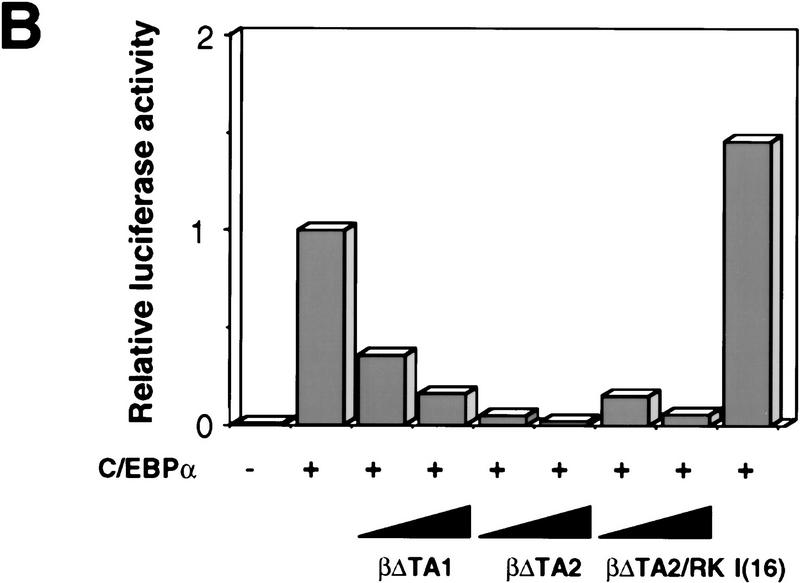
Figure 5.
Characterization of dominant negative mutants of C/EBPβ. (A) Structure of dominant-negative rat C/EBPβ mutants. (*) Position of the RK mutation. The sequences show a comparison between the basic regions (underlined) of rat C/EBPα and C/EBPβ, and the position of the substitutions introduced in the C/EBPβ/RK mutant (arginine-238 and lysine-239 into alanines). The numbers indicate the positions of the first and last amino acid shown for each protein. (B) Capacity of mutant C/EBPs to block the transactivation of the mim-1 promoter by C/EBPα. The ability of the βΔTA1, βΔTA2, and βΔTA2/RK alleles to inhibit C/EBP dependent transactivation was tested on the mim-1 promoter. One microgram of pMim-1ΔC–LUC was cotransfected with 0.25 μg of pRSVβGal, 0.5 μg pCMV–C/EBPα (chicken C/EBPα expressed from the CMV promoter). Two and five micrograms of CMV expression vector for the dominant-negative alleles were cotransfected as indicated. As a specificity control, 5 μg of a CMV expression vector for the I(16) protein (a dominant-negative inhibitor of the AML complex) was used. Relative transactivation values were calculated as in Fig. 3; the activity of pMim-1ΔC–LUC in the presence of C/EBPα was arbitrarily assigned a value of 1.