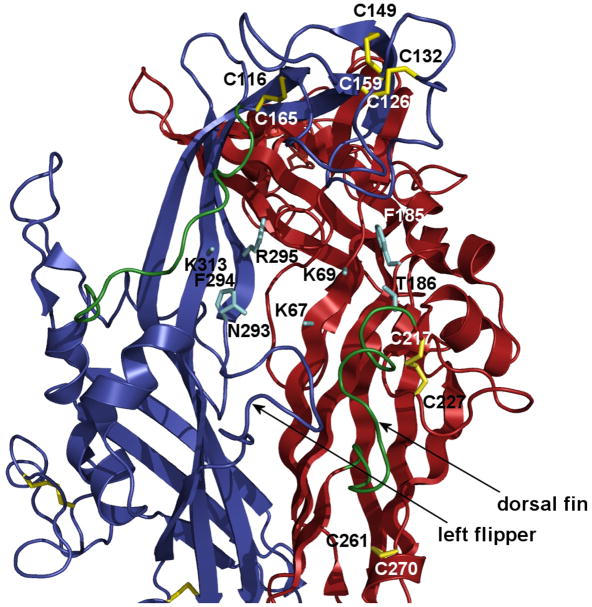
Fig. 4.
Model of two zP2X4.1R subunits with the putative ATP binding site (cyan) and position of conserved disulfide bonds (yellow) based on the crystal structure data (Kawate et al. 2009). The following conserved residues (rat P2X4R numbering) are predicted to form ATP the binding site at the interface between two subunits: K67, K69, F185 and T186 in the red subunit and N293, F294, R295 and K313 in the blue subunit (Ennion et al. 2000; Jiang et al. 2000; Roberts and Evans 2004, 2006; Yan et al. 2006; Zemkova et al. 2007) . The third subunit was removed for clarity but would be behind the two shown. Two turns (green) might be relaxed by disruption of the SS1 and SS4 bonds. The turn that precedes the C217 residue is termed the dorsal fin and is facing the left flipper (blue turn). This interface may involve interaction between subunits because earlier experiments showed that alanine substitution of single residue in the left flipper of the P2X3R (aspartate 280, the rat P2X4 numbering) significantly changes kinetics of receptor resensitization (Fabbretti et al. 2004).