Figure 6.
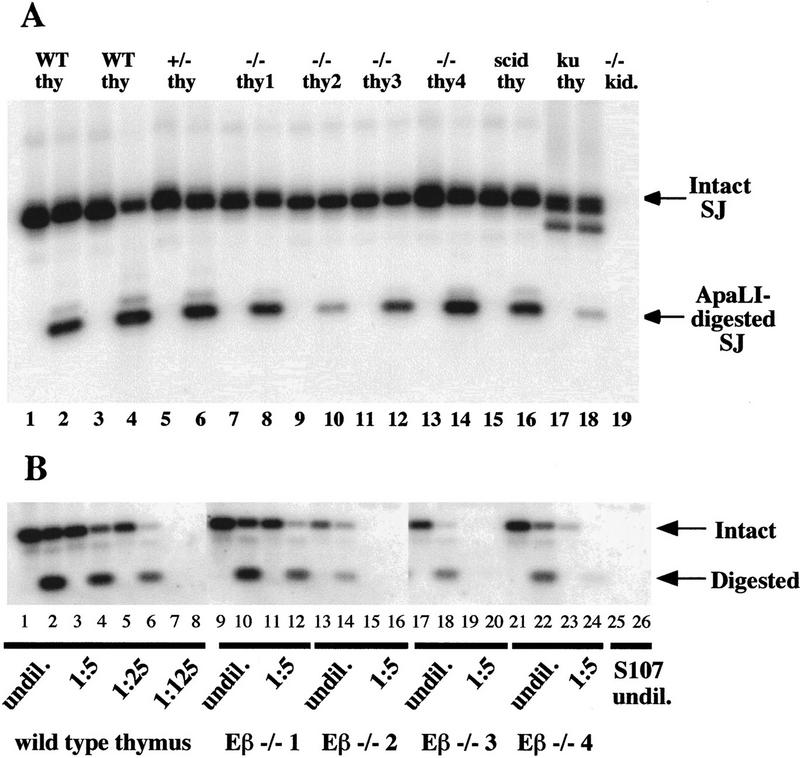
Broken TCRβ SEs are resolved into SJs in Eβ−/− thymocytes. SJ formation involving a 3′ of Dβ RSS and a 5′ of Jβ RSS results in an extrachromosomal circular product of characteristic structure. Each broken SE generated by recombinase cleavage is blunt and shows no loss or gain of nucleotides from the conserved RSS. Therefore, fusion of these ends generates a new site for the enzyme ApaLI (5′-GTGCAC-3′). Dβ-to-Jβ SJs of this structure can be detected in a pool of total genomic DNA by PCR amplification with primers on either side of the junction and hybridization with an internal oligonucleotide probe. Precision of amplified signal joints can be confirmed by ApaLI digestion. Digestion will result in two products of defined size, one of which will hybridize to the probe. (A) SJs involving Dβ2 and Jβ2.6 RSSs were amplified from genomic DNA by use of a scheme similar to that described above, except that nested primers were used in a two-step PCR reaction. Following amplification, one-half of each PCR product was digested with ApaLI at 37°C (even numbered lanes) and the other half was mock digested with a heat-inactivated irrelevant restriction enzyme on ice (odd numbered lanes). The reaction products were then resolved on an agarose gel, Southern blotted, and hybridized with an oligonucleotide probe that will detect the undigested SJ (intact SJ) as well as one of the two fragments generated by ApaLI cleavage of the SJ (ApaLI-digested SJ). Reactions contained template genomic DNA from two independent wild-type thymuses (lanes 1-4), an Eβ+/− thymus (lanes 5, 6), four individual Eβ−/− thymuses (lanes 7–14), a scid thymus (lanes 15,16), a Ku86−/− thymus (lanes 17, 18) or Eβ−/− kidney (lane 19). (B) Quantitation of Dβ-to-Jβ SJs. Serial five-fold dilutions of genomic DNA from a wild-type thymus and four independent Eβ−/− thymuses were made into genomic DNA from the S107 plasmacytoma cell line. Extrachromosomal SJs involving the Dβ2 and Jβ2.6 RSSs were amplified from each sample using a two-step nested PCR reaction exactly as in A. One-half of each PCR product was digested with ApaLI (even numbered lanes) and the other half was mock digested (odd numbered lanes). The source of DNA and its dilution factor are indicated below each pair of lanes. The intact and digested products of the expected sizes are indicated by arrows. S107 cells do not rearrange TCRβ genes and therefore do not contain detectable TCRβ signal joints (lanes 25,26).
