Abstract
ENT1 of Arabidopsis thaliana was the first member of the equilibrative nucleoside transporter (ENT) family to be identified in plants and characterized as a cellular, high-affinity nucleoside importer. Evidence is presented here for a tonoplast localization of ENT1 based on proteome data and Western blot analyses. Increased export of adenosine from reconstituted tonoplast preparations from 35S:ENT1 mutants compared with those from the wild type and ENT1-RNAi mutants support this view. Furthermore, increased vacuolar adenosine and vacuolar 2′3′-cAMP (an intermediate of RNA catabolism) contents in ENT1-RNAi mutants, but decreased contents of these metabolites in 35S:ENT1 over-expresser mutants, were observed. An up-regulation of the salvage pathway was detected in the latter mutants, leading to the conclusion that draining the vacuolar adenosine storage by ENT1 over-expression interferes with cellular nucleotide metabolism. As a consequence of the observed metabolic alterations 35S:ENT1 over-expresser mutants exhibited a smaller phenotypic appearance compared with wild-type plants. In addition, ENT1:RNAi mutants exhibited significantly lower in vitro germination of pollen and contained reduced internal and external ATP levels. This indicates that ENT1-mediated nucleosides, especially adenosine transport, is important for nucleotide metabolism, thus influencing growth and pollen germination.
Keywords: Arabidopsis thaliana L., equilibrative nucleoside transporter, pollen, purine, pyrimidine, salvage pathway, vacuole
Introduction
Nucleotides are ubiquitous in metabolism: in addition to being components of DNA and RNA, and the currency of energy metabolism in the cell, they also are formed into essential coenzymes, secondary metabolites, and plant hormones (reviewed in Zrenner et al., 2006).
Nucleotide metabolism is divided in de novo synthesis, salvage reactions, and degradation. During nucleotide de novo synthesis, nitrogen in the form of amino acids and energy in hte form of ATP are consumed. The proposed function of the salvage pathway is the recycling and thus preservation of nitrogen and energy within the purine or pyrimidine basic structure.
The metabolic intermediates which can be salvaged are the nucleobases and nucleosides. Both can also be exchanged between cells by specialized transport proteins. The nucleoside transport proteins in Arabidopsis belong to the equilibrative nucleoside transporter (ENT) family. Members of this protein family mediate the distribution of purine and pyrimidine nucleosides (Möhlmann et al., 2001; Wormit et al., 2004) and this carrier group comprises eight members. Five of them, ENT1, 3, 4, 6, and 7 have so far been characterized at the biochemical level by heterologous expression in baker's yeast (Möhlmann et al., 2001; Li et al., 2003; Wormit et al., 2004; Chen et al., 2006). All of these carriers exhibit broad substrate specificity and transport purine and pyrimidine nucleosides with apparent high affinities ranging from 3 μM to 90 μM. It was shown recently, that a strict regulation of nucleoside degradation at the level of nucleoside hydrolase is essential to balance nitrogen pools within nucleotides and amino acids (Jung et al., 2009, 2011). This is especially true for the early stages of seedling germination and, subsequently, when expression of the main nucleoside hydrolase activity (NSH1) is high in roots (Jung et al., 2009, 2011). Nucleobases generated by this process can be exchanged between cells by the corresponding transporters that exist in Arabidopsis in large numbers and belong to several protein families (Möhlmann et al., 2010). Recently, two nucleobase transporters were identified (AtAzg1 and AtAzg2), which show substantial amino acid sequence similarity to the adenine-guanine-hypoxanthine transporter AzgA of Aspergillus nidulans (Mansfield et al., 2009). Analysis of single and double mutant lines harbouring the T-DNA insertion alleles AtAzg1-1 and AtAzg2-1 revealed a marked resistance to growth in the presence of 8-azaadenine and 8-azaguanine, substantiating a purine base transport function of these proteins in planta (Mansfield et al., 2009). Purine salvage is best analysed at the level of adenosine kinase (AK). Two isoforms have been identified, located in the cytosol (Moffatt et al., 2002). Plants lacking AK activity show a reduced capacity to perform methylation reactions underlining the relative importance of these enzyme reactions (Moffatt et al., 2002). The salvage of pyrimidine nucleosides and nucleobases requires the activity of uridine kinase (UK) and uracil phosphoribosyltransferase (UPRT), respectively. At5g40870 (AtUK/UPRT1) was characterized as a tandem enzyme exhibiting both UK and UPRT activity (Islam et al., 2007). However, this result was questioned by others, claiming that At3g53900 (UPP) is responsible for almost all UPRT activity in Arabidopsis (Mainguet et al., 2009). Whereas AtUK/UPRT1 is located in the cytosol, UPP is a plastid-located enzyme (Islam et al., 2007; Mainguet et al., 2009). Four further genes exist in Arabidopsis containing a UK and a UPRT domain, their exact functions, however, so far remains elusive.
Uptake of nucleosides into seedlings almost exclusively occurs via ENT3 (Chen et al., 2006; Traub et al., 2007). However, quantitative RT-PCR clearly showed that seedlings also express ENT1 at high levels in addition to ENT3. Therefore, a question was asked about the specific contribution of each of these ENTs in nucleoside uptake and further degradation or salvage. Furthermore, the previously reported localization of ENT1 at the plasma membrane (Li et al., 2003; Wormit et al., 2003) contradicts the finding that ENT1 obviously does not contribute significantly to nucleoside transport at the plasma membrane level (Traub et al., 2007). To resolve this discrepancy, the tissue-specific expression was analysed and the subcellular localization of ENT1 was critically reinvestigated. So far, nucleotide metabolism in plant vacuoles has not been studied. However, the presence of RNA oligonucleotides in vacuoles from tomato cell culture and the presence of RNAse activity in vacuoles indicate that RNA degradation may occur in this storage compartment. (Abel et al., 1990; Abel and Glund, 1992; Löffler et al., 1992). In yeast (Saccaromyces cerevisiae) an ENT homologue, FUN26 is known to reside in endosomal membranes and it is speculated that this transporter is involved in the release of RNA breakdown products from these vesicles (Vickers et al., 2000).
Pollen represents an interesting object to study purine and pyrimidine metabolism. Besides transport of pyrimidine and purine nucleosides (Kamboj and Jackson, 1985, 1987) nucleotide salvage was shown to be of crucial importance for pollen development. This was shown, for example, by Arabidopsis mutants lacking the major isoform of pollen adenine phosphoribosyl transferase (APRT) who do not reach fertility (Moffatt and Sommerville, 1988). Interestingly, a mutant lacking PUR4, encoding the fourth enzymatic step in purine de novo synthesis was also lethal to the male gametophyte (Berthome et al., 2008). Theoretically, purine de novo synthesis and salvage should be complementary to each other, given that nucleosides or bases can be imported by pollen. However, a complementation at this step obviously does not occur.
In the last few years, extracellular ATP (eATP) has been established as a signalling molecule in plants. eATP may enter the apoplast through secretory vesicles or by specialized ABC-type transporters (Roux and Steinebrunner, 2007) and is supposed to bind to ATP receptors, similar to the situation in animals (Burnstock, 2009). Two types of plant tissues for which this type of signalling has been demonstrated are roots and pollen (Kim et al., 2006; Wu et al., 2007). In addition to export, apyrases which cleave ATP play an important role in maintaining optimal eATP concentrations for growth. Mutants lacking both apyrase isoforms showed a dwarf phenotype due to reduced cell elongation (Wu et al., 2007). According to a current model, eATP concentrations support growth at an optimal concentration, whereas higher and lower concentrations inhibit growth (Roux and Steinebrunner, 2007).
In this work, the aim was to unravel the physiological function of ENT1, critically reinvestigating the subcellular localization of this protein.
Materials and methods
Plant material and growth
Wild-type and transgenic Arabidopsis thaliana (L.) Heynh. plants (ecotype Columbia) were used throughout the present studies. Plant growth was carried out at 22 °C and 120 μmol quanta m−2 s−1 in a 10/14 h light/dark regime in standardized potting soil (ED73, Einheitserde u. Humuswerke GmbH, Sinntal/Jossa, Germany: www.einheitserde.de).
Isolation of vacuoles and reconstitution
Intact vacuoles were isolated from 35-d-old Arabidopsis leaves according to Robert et al. (2007). For this, 6 g of leaf material was harvested at the end of the night and enzymatically digested. Protoplasts formed were washed, lysed, and subjected to ultracentrifugation on a Ficoll gradient. Activity of α-mannosidase as a vacuolar marker was determined spectrophotometrically and used as a reference for all further analyses.
Vacuolar membranes were prepared from vacuoles after freezing, thawing, and ultracentrifugation (100 000 g, 60 min). 7 μg of membrane protein were reconstituted into liposomes prepared from soybean phosphatidylcholine (100 mg ml−1) as given in Möhlman et al. (1995). Liposomes were preloaded with 50 mM [14C]-adenosine and resuspended in buffer at a pH of 7.5 after passage through NAP-10 (GE-Healthcare) columns.
Quantitative RT-PCR
Quantitative RT-PCR was performed as given in Leroch et al. (2005). Gene-specific oligonucleotides used are listed in Supplementary Table S1 at JXB online. Elongation factor 1α was used as the reference gene.
Generation of mutants and histochemical localization of GUS activity
To generate transgenic RNAi plants, a 479 bp fragment from ENT1 (corresponding to bp 32–511) was cloned in sense and antisense orientations into the pHANNIBAL vector (Wesley et al., 2001). The primers used are listed in Supplementary Table S1 at JXB online. To generate ENT1 over-expression mutants, the complete coding region of ENT1 (At1g70330) was inserted into the pHANNIBAL vector (Wesley et al., 2001) via XbaI and XhoI restriction sites. The resulting pHANNIBAL constructs were subcloned into the binary vector pART27 as given in Reiser et al. (2004). Subsequently, the plasmids were used for Agrobacterium tumefaciens transformation. Transformation of Arabidopsis was performed according to the floral-dip method (Clough and Bent, 1998).
For the generation of the promoter-GUS-constructs the binary vector pGPTV (Becker et al., 1992) was used. For the generation of the ENT1-promoter–GUS fusions a promoter region of about 1.1 kbp was cloned upstream of the GUS gene, respectively. For amplification of the promoter region, the primers used are listed in Supplementary Table S1 at JXB online. After ligation of the PCR product with the GUS-gene, the resulting construct was used for Agrobacterium and Arabidopsis transformation (see above). From each construct more than 10 independent lines were analysed and representative results were presented. For root sections, 6-d-old plants were prefixed in 50 mM phosphate buffer, pH 7.2, containing 1.4% formaldehyde (Schmidt et al., 2004). Samples were stained according to standard protocols (Weigel and Glazebrook, 2002).
Isolation of Arabidopsis pollen and germination experiments
Freshly harvested flowers from 24 plants were vigorously shaken in a capped centrifugation tube with 10 ml H2Obidest. Subsequently, the released pollen were filtered through a 250 μm mesh and pelleted by centrifugation (1000 g, 1 min). The pellet was resuspended in 50 mM K2HPO4, pH 5.8, 1% (w/v) sucrose, and 4% (w/v) sorbitol, and the pollen grains were counted in a Neubauer chamber.
In vitro pollen germination was performed according to Fan et al. (2001) on germination agar plates consisting of 5 mM MES (2-(N-morpholino) ethanesulphonic acid), pH 5.8, 1 mM KCl, 10 mM CaCl2,.8 mM MgSO4 0, 1.5 mM H3BO3, 16.6% (w/v) sucrose, 3.65% (w/v) sorbitol, 10 μg ml−1 myo-inositol, and 1% (w/v) agar. Freshly opened flowers were collected from several plants at random and dipped onto the agar surface to release pollen grains. Pollen germination was determined by microscopy after 16 h incubation in a chamber at 25 °C and 100% relative humidity in the dark. On each plate 1000 pollen grains were counted for calculation of the pollen germination rate.
Uptake experiments on pollen
To measure adenosine uptake, pollen was isolated as described above and resuspended in medium A [50 mM K2HPO4, pH 5.8, 16% (w/v) sucrose, and 4% (w/v) sorbitol]. To initiate uptake, 100 μl of pollen suspension (106 pollen ml−1) were added to 100 μl medium A containing 4 μM [2,8-3H] adenosine (50 GBq mmol−1). Pollen grains were incubated for the indicated time at 25 °C with gentle shaking and subsequently filtered on membrane filters (0.45 μm, Whatman, Clifton, NJ, USA). After washing three times with buffer medium A, filters were transferred to scintillation tubes and radioactivity on the filters was quantified by liquid-scintillation counting in a Tricarb 2500 TR scintillation counter (Packard, Dreieich, Germany).
Quantification of extracellular ATP
To measure the amount of ATP exported by the pollen grains, the rLuciferase/Liciferin Reagent (Promega, USA) was used the ENLITEN according to the manufacturer's advice. Test tubes were prepared containing 1 ml pollen germination agar. To start the incubation, 10 μl of pollen suspension prepared as described above was dripped on to the surface of the agar.
After incubation for the indicated time 400 μl 50 mM TRIS buffer medium (pH 8) and 100 μl ENLITEN rLuciferase/Liciferin Reagent were added and luminescence was quantified after 1 min in a BioOrbit 1253 Luminometer (Bio Orbit, Finnland).
Intracellular ATP content in pollen grains
100 μl of pollen suspension prepared as described above were mixed with 400 μl of (7% (v/v) HClO4 and 10 mM EDTA) and incubated for 15 min on ice. Afterwards, a mixture of 5 M KOH and 1 M triethanolamine were added until pH 8.0 was reached and incubated for a further 10 min on ice prior to centrifugation for 5 min at 10 000 g. 10 μl supernatant were mixed with 400 μl 50 mM TRIS buffer medium (pH 8) and 100 μl assay medium and luminescence was quantified in a BioOrbit 1253 Luminometer (Bio Orbit, Finnland).
Quantification of adenine derivatives by HPLC
Plant leaf tissue was harvested, immediately frozen, and thenground in liquid nitrogen. Each sample of 100 mg was mixed with 500 μl ethanol (80% v/v) and incubated for 20 min at 80 °C in a shaker. After centrifugation for 5 min at 15 000 g, the supernatant was collected and ethanol was removed in a vacuum centrifuge (RC 10-10, Jouan, Saint Nazaire, France). The dried pellet was resuspended in 250 μl distilled water. To sensitize the measurement of adenine derivatives, samples were derivatized to 1,N6-ethenoadenine. For this purpose 170 μl sample was mixed with 40 μl buffer (330 mM citrate and 380 mM KH2PO4, pH 5.2) and 40 μl chloracetaldehyde solution (50% w/v) and incubated for 15 min at 80 °C. HPLC analysis was carried out with a DIONEX system consisting of a P 680 HPLC pump; an ASI-100 Automated Sample Injector; an RF 2000 Fluorescence Detector; a UCI-50 Universal Chromatography Interface (Dionex, Sunnyvale, USA); and a Nucleodur 100-5 C18 ec column (Machery and Nagel, Düren, Germany). As the eluent, a solution of 10 mM KH2PO4 pH 5.4 and 5.7 mM tetrabutylammonium hydrogen sulphate with gradually increasing (0–80%) acetonitrile, was used.
Results
Tissue-specific expression and subcellular localization of equilibrative nucleoside transporters (ENTs) in Arabidopsis
A starting point for the analysis presented here was the identification of ENT3 as the sole pyrimidine nucleoside uptake system in Arabidopsis seedlings (Chen et al., 2006; Traub et al., 2007). Therefore, a question was asked about the physiological function of other ENT-type transporters at this developmental state. Quantitative RT-PCR analysis revealed that besides ENT3- only ENT1-transcripts accumulated markedly in seedlings (see Supplementary Fig. S1 at JXB online). ENT1 transcript levels were 85-fold higher and ENT3 transcript levels were 25-fold higher compared with those of ENT8, which was in third place (see Supplementary Fig. S1 at JXB online). A more detailed analysis of ENT1 tissue-specific expression by corresponding promoter– reporter gene fusions was followed by a critical re-investigation of the subcellular localization of ENT1, so far assumed to reside in the plasma membrane (Li and Wang, 2000; Wormit et al., 2004).
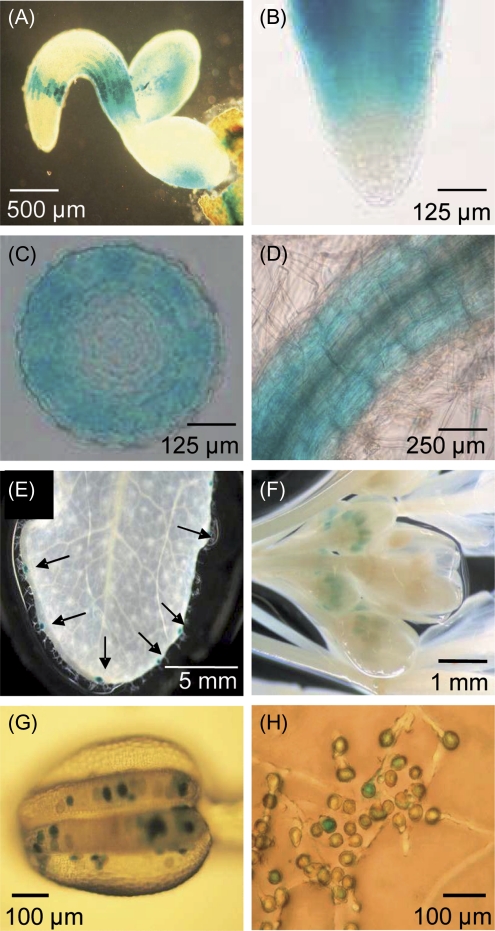
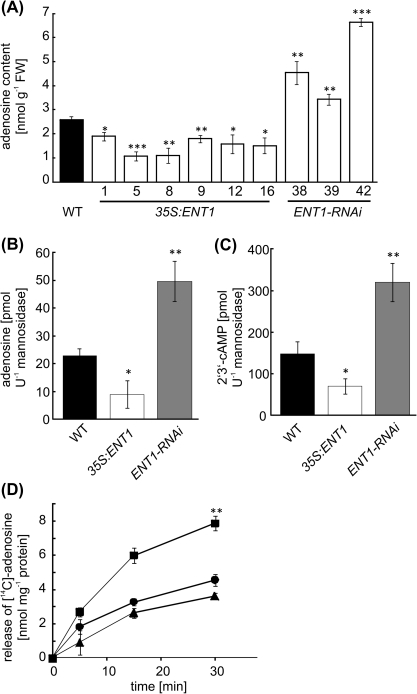
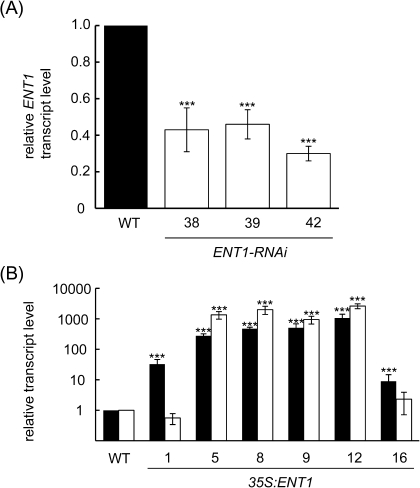
Seedlings carrying ENT1:GUS constructs that were analysed 2 d after germination had staining in distinct regions of the root; within the elongation zone, in the root-hair , and at the transition to the shoot (Fig. 1A, B). In cross-sections, most of the staining was confined to the root cortex (Fig. 1C, D). Cotyledons from ENT1:GUS seedlings were also characterized by an intense accumulation of colour (Fig. 1A). In fully developed leaves, GUS staining was restricted to hydathodes (Fig. 1E). In mature flowers, GUS expression was restricted to pollen (Fig. 1F, G). In vitro-germinated pollen show GUS expression in mature pollen tubes, whereas in germinated pollen tubes no staining was visible (Fig. 1H). In a proteome study, ENT1 was found in the vacuolar membrane of Arabidopsis mesophyll cells (Jaquinod et al., 2007) and this was confirmed with our own proteome studies (O Trentmann and HE Neuhaus, unpublished data). To verify this result, highly purified vacuoles (see Supplementary Fig. S2A at JXB online) were probed with an ENT1-specific polyclonal antiserum which stained a protein of the expected size of 49 kDa (see Supplementary Fig. S2B at JXB online). By contrast, in a crude extract, no staining was visible. Similarly, the preimmune serum showed no detection of protein (see Supplementary Fig. S2C at JXB online). To corroborate these findings, ENT1 mutants were analysed for their adenosine contents, which represents the main substrate of ENT1. For this, an RNAi-approach was chosen and, in addition, 35S:ENT1 mutants were generated. While transcript levels were reduced to 46%, 43%, and 30% in seedlings of ENT1-RNAi-lines #39, #38, and #42, respectively (Fig. 2A) in six over-expresser lines (1, 5, 8, 9, 12, and 16) the ENT1 transcript level was elevated 9-fold to 1060-fold (Fig. 2B). When the adenosine contents in seedlings of both types of mutants were analysed, it became obvious that ENT1-RNAi-lines contained more adenosine compared with the WT, whereas 35S:ENT1 lines contained less adenosine (Fig. 3A). In detail, the adenosine content of WT leaves was 2.6 nmol g−1 FW, whereas over-expresser mutants contained 1.1–1.9 nmol adenosine g−1 FW (Fig. 3A). By contrast, the adenosine contents of ENT1-RNAi mutants were increased relative to WT leaves and accounted for 3.4–6.6 nmol g−1 FW (Fig. 3A). Assuming that ENT1 resides in the vacuolar membrane, it was of interest to check whether the main substrates of ENT1, adenosine, could be found inside the vacuole. Therefore, a lumen fraction of highly purified vacuoles was concentrated and derivatized with chloroacetaldehyde. Subsequently, adenine derivatives were analysed by HPLC. Besides significant amounts of adenosine, 2′3′-cyclic AMP, an intermediate of RNA degradation was the only further derivatized metabolite detected in the analysis. Measured contents of adenosine in vacuoles prepared from WT, 35S:ENT1 line #9 and ENT1-RNAi line #42 were 22.80±2.66 pmol U−1mannosidase, 9.1±4.84 pmol U−1 mannosidase, and 49.6±7.23 pmol U−1 mannosidase, respectively (Fig. 3B). In addition, 2'3'-cyclic AMP was quantified and accounted for 147.8±29.54 pmol U−1 mannosidase in WT vacuoles, 70.2±18.32 pmol U−1 mannosidase in 35S:ENT1 (line #9) vacuoles, and 320.2±45.89 pmol U−1 mannosidase in ENT1-RNAi (line #42) vacuoles, respectively (Fig. 3C).
Fig. 1.
Expression analysis of ENT1 by the use of corresponding promoter–GUS lines. (A–C) Two-d-old seedlings. (A) Whole seedling, (B) root tip, (C) cross-section of root elongation zone, (D) 10-d-old seedling, root hair zone; (E) developed leaf, arrows showing stained hydathodes, (F) flower, (G) anther with pollen, (H) germinated pollen.
Fig. 2.
Contents of adenosine and 2′3′-cAMP in leaves and isolated vacuoles of ENT1 mutants and release of adenosine from liposomes reconstituted with vacuolar membrane protein. (A) Adenosine contents in leaves from WT, 35S:ENT1 overexpresser lines, and ENT1-RNAi lines; (B) adenosine contents in isolated vacuoles from WT, 35S:ENT1 over-expresser line #9, and ENT1-RNAi line #42; (C) 2'3'-cAMP contents in isolated vacuoles from WT, 35S:ENT1 over-expresser line #9, and ENT1-RNAi line #42; (D) release of [14C]-adenosine from preloaded liposomes reconstituted with vacuolar membrane protein from WT (circles), 35S:ENT1 over-expresser line #9 (squares), and ENT1-RNAi line #38 (triangles). All data shown represent the mean of at least three independent experiments (6SE). The asterisks indicate significant differences between WT and mutants, based on Student's t test (*P <0.05, **P <0.01, ***P <0.005).
Fig. 3.
Relative transcript levels of ENT1 mutants. Whole seedlings were used for RNA extraction. (A) ENT1-RNAi lines were analysed for ENT1 transcript accumulation by quantitative RTPCR. (B) 35S:ENT1 over-expresser lines were analysed for ENT1 (black bars) and ENT7 (white bars) transcript accumulation by quantitative RT-PCR. (A, B) WT transcript levels were set to one. Data represent the mean of at least three independent experiments (6SE). The asterisks indicate significant differences between WT and mutants, based on Student's t test (*P <0.05, **P <0.01, ***P <0.005).
These results point to a function of ENT1 in the release of adenosine from the vacuole. To verify this assumption experimentally, vacuolar membrane protein was reconstituted into liposomes preloaded with [14C]-adenosine (50 mM), and the release of label was monitored over 30 min. While liposomes containing vacuolar membrane protein from the WT released a maximum amount of label after 30 min incubation, accounting for 4.56 nmol mg−1 protein, liposomes containing vacuolar membrane protein from 35S:ENT1 over-expresser line #9 released double the amount compared with WT liposomes, a maximum of 7.87 nmol mg−1 protein after 30 min (Fig. 3D). Liposomes containing vacuolar membrane protein from ENT1-RNAi line #38 released slightly less adenosine compared with wild-type controls at all three time points measured. By applying a similar reconstitution technique it was shown that only membrane protein inserted in the native orientation were functional (Trentmann et al., 2007) supporting the view that ENT1 functions in adenosine export from the vacuole in vivo.
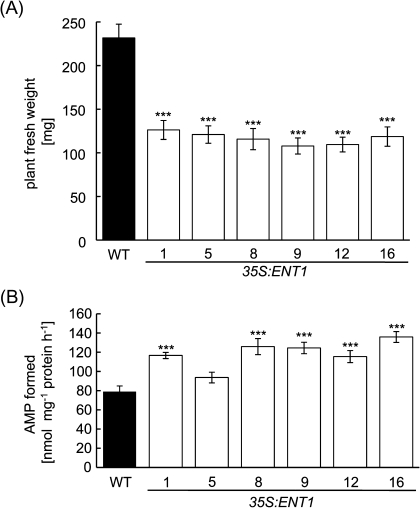
In addition to these metabolic alterations, all 35S:ENT1 over-expressers showed clear growth deficiencies on soil 5 weeks after germination (Fig. 4A). Analysis of fresh weights of above-ground tissues from such plants revealed that ENT1-over-expression lines accumulated 107.9–126.7 mg fresh weight while WT plants accumulated 231.7 mg fresh weight (Fig. 4A). These growth deficiencies were accompanied by an increased activity of the salvage pathway enzyme adenosine kinase (AK). AK activity increased by 20–40% in most 35S:ENT1 lines, except for line #5 which exhibited an increase in AK activity of 10% (Fig. 4B). The AK activity of wild-type leaves accounted for 80 nmol mg−1 protein h−1 (Fig. 4B). Furthermore, a marked increase in ENT7 transcript level was noted whereas all other ENT transcripts were unaffected. In seedlings, this increase was dramatic and quite comparable with the increase in ENT1 transcript accumulation in most mutant lines (Fig. 2B).
Fig. 4.
Characterization of 35S:ENT1 lines. (A) Fresh weights of 5-week-old WT and 35S:ENT1 mutant plants grown under short-day conditions. 15 plants per line were analysed, and similar results were obtained in at least two further, independent cultivation experiments. (B) Activity of adenosine kinase in seedlings from WT and 35S:ENT1 mutant plant lines. Data represent the mean of three independent experiments (±SE). The asterisks indicate significant differences between mutant and WT samples, based on Student's t test (* P<0.05, **P <0.01, ***P <0.005).
Molecular analysis of ENT1 over-expressers and RNAi-mutants with respect to pollen morphology and physiology
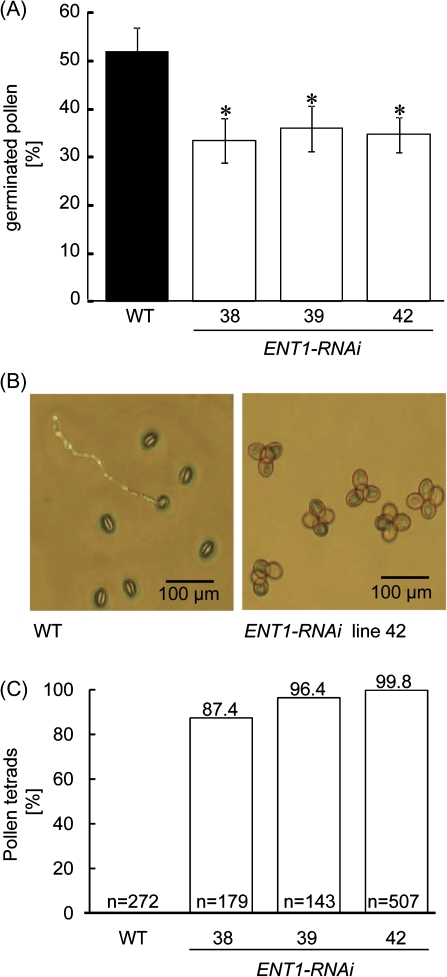
As ENT1 showed marked expression in pollen (Fig. 1F–H), it was investigated whether corresponding mutants interfere with pollen development or germination. In the ENT1-RNAi mutants in vitro germination of pollen on agar medium according to Fan et al. (2001) was significantly reduced and pollen appeared in the form of tetrads (Fig. 5), whereas 35S:ENT1 over-expresser mutants were unobtrusive. Therefore, we concentrate on the analysis of ENT1-RNAi lines in the following. Whereas WT pollen germinated to about 52% after 20 h incubation on synthetic germination medium, RNAi mutants exhibited only 33.5%, 36%, and 34.7% germination efficiency in lines 38, 39, and 42, respectively (Fig. 5A). It was observed that pollen from all RNAi mutants appear as tetrads when spotted directly on to agar whereas pollen from wild-type plants produce single pollen (Fig. 5B). 87.4–99.8% of RNAi mutant pollen showed tetrad formation, whereas this was never observed in WT pollen (Fig. 5C).
Fig. 5.
Analysis of pollen morphology and in vitro germination. (A) Germination efficiency of WT and ENT1-RNAi mutant pollen. Freshly opened flowers were dipped on agar plates and germination was monitored after 16 h of incubation. Data represent the mean of at least six independent experiments (±SE). The asterisks indicate significant differences, based on Student's t test (*P <0.05). (B) Representative images of WT and ENT1-RNAi mutant pollen. (C) Abundance of mature pollen appearing as tetrads. Single pollen and tetrads were counted after dipping anthers onto agar plates. Standard errors are below 5%. (This figure is available in colour at JXB online.)
Adenosine uptake and contents of internal and extracellular ATP in ENT1-RNAi mutant pollen
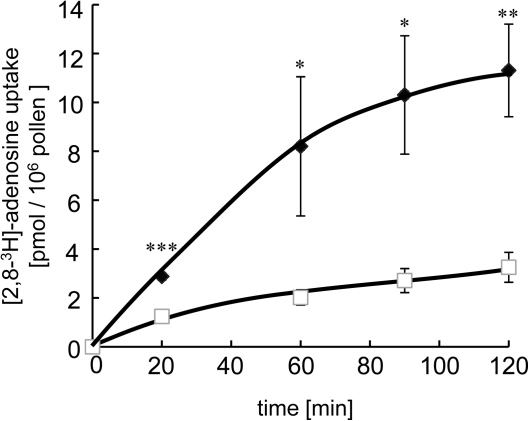
Nucleoside uptake had been demonstrated before on petunia pollen (Kamboj and Jackson, 1995). To test whether Arabidopsis pollen grains are also able to take up nucleosides, the import of radiolabelled adenosine was measured. [2,8-3H]-labelled adenosine could be imported into intact pollen, showing a constant rate for at least 20 min. After 20 min, uptake accounted for 3 pmol/106 pollen (Fig. 6). After this time, adenosine import further increased up to 120 min (Fig. 6). Interestingly, uptake of adenosine in the strongest RNAi-line #42 was markedly reduced at all the time points analysed. After 60 min WT pollen exhibited an adenosine import of 8 pmol/106 pollen whereas RNAi line #42 only imported 1 pmol/106 pollen (Fig. 6).
Fig. 6.
Adenosine uptake in pollen from WT- and ENT1-RNAi-mutant plants. Time-dependent uptake of [2,8-3H]-adenosine [3 μM] into freshly isolated pollen from WT (open squares) and ENT1-RNAi line #42 (closed diamonds). The asterisks indicate significant differences between mutant and WT samples based on Student's t test (*P <0.05, **P <0.01, ***P <0.005).
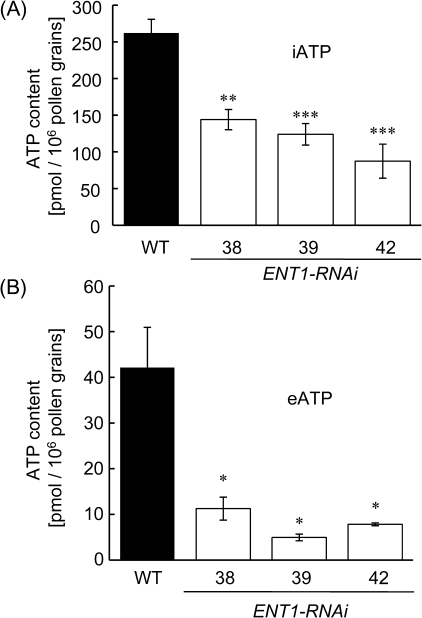
The reduced ENT1 transcript levels of ENT1-RNAi lines were accompanied by markedly lowered internal ATP contents (Fig. 7A). Internal ATP (iATP) was measured with a luciferin/luciferase system on perchloric acid extracts of pollen. In WT pollen, 262 pmol ATP/106 pollen were detected, whereas ENT1-RNAi lines #38, #39, and #42 contained only 144 pmol ATP/106 pollen, 124 pmol ATP/106 pollen, and 87 pmol ATP/106 pollen, respectively (Fig. 7A).
Fig. 7.
Levels of intracellular and extracellular ATP of pollen from WT and ENT1:RNAi mutants. ATP was quantified by a coupled luciferin/luciferase assay. (A) Intracellular ATP (iATP), extracted with perchloric acid from pollen, (B) extracellular ATP (eATP) was determined after pollen were incubated on pollen germination agar for 30 min. Black bars, WT; white bars, ENT1-RNAi mutants. Data represent the mean of three independent experiments (±SE) The asterisks indicate significant differences between mutant and WT samples based on Student's t test (*P <0.05, **P <0.01, ***P <0.005).
In recent years a role of extracellular ATP in plant growth processes became obvious (Demidchik et al., 2003; Jeter et al., 2004) and this also holds true for pollen germination as demonstrated by inhibitory effects of exogenously added ATP and by studies with apyrase knockout mutants (Steinebrunner et al., 2003; Wolf et al., 2007). Therefore, the ATP export of a pollen suspension spread on to agar was determined with a luciferase/luciferin test system. First, in a time-dependent assay, ATP export was observed linearly for 20 min. At this time point, WT pollen released 42.1 pmol ATP/106 pollen (Fig. 7B). ENT1-RNAi-lines #38, #39, and #42 released only 11.3 pmol ATP/106 pollen, 5.0 pmol ATP/106 pollen, and 7.8 pmol ATP/106 pollen, respectively (Fig. 7B). These data show that limiting the level of ENT1 correlates with a decrease in ATP export from pollen grains leading to lowered eATP levels.
Discussion
ENT1 represents a tonoplast-localized nucleoside transporter
By the marked resistance of ENT3:T-DNA mutants towards 5-fluoro-uridine, it can be concluded that no other nucleoside transporter is active as a cellular importer for uridine in Arabidopsis seedlings (Chen et al., 2006; Traub et al., 2007). This is interesting in so far as ENT1 and ENT3 are expressed at similar levels in seedlings (see Supplementary Fig. S1 at JXB online). Therefore, the question arises why ENT1 cannot compensate for the loss of ENT3. According to biochemical analysis, ENT1 is, like ENT3, most probably a nucleoside-proton symporter and able to perform adenosine as well as uridine import (Möhlmann et al., 2001; Chen et al., 2006). This means that ENT1 should be able to take over the function of ENT3 in corresponding mutants. Furthermore, by the use of GFP-fusion proteins, ENT1 was shown to reside in the plasma membrane (Li and Wang, 2000; Möhlmann et al., 2001). However, these results contradict the identification of ENT3 as the sole nucleoside uptake system in seedings. In a proteome study, ENT1 was found in the vacuolar membrane of Arabidopsis mesophyll cells (Jaquinod et al., 2007) and this was confirmed with our own proteome studies (O Trentmann and HE Neuhaus, unpublished data). Such a subcellular localization, however, would explain the discrepancies discussed before. As a proton symporter, ENT1 would function as a nucleoside exporter from the vacuole, leading to the question about the physiological function of such a vacuolar nucleoside transport activity. It is reported that vacuoles from tomato contain nucleosides and further RNA breakdown products (Leinhos et al., 1986; Abel et al., 1990). Therefore, one might speculate that RNA degradation, in part, takes place in the vacuole and ENT1 is involved in the export of liberated nucleosides for salvage or for final breakdown.
This view is further strengthened by the identification of a vacuolar RNase isoform (Abel and Glund, 1986; Löffler et al., 1992). In addition to the proteome data and the ENT3:T-DNA mutant resistance towards 5-fluoro-uridine, further evidence for a localization of ENT1 in the vacuolar membrane could be gathered by (i) identification of an acid-di-leucine motive in the N-terminus of ENT1 (amino acid positions 41–46) which is known to act as a sorting signal to the vacuole in plants (Yamada et al., 2010), (ii) detection of ENT1 in vacuolar membranes by a polyclonal antiserum, (iii) higher total adenosine contents in ENT1-RNAi mutants compared with WT plants and, in contrast, lower adenosine contents in 35S:ENT1 over-expresser mutants, and (iv) lower adenosine and 2'3'-cAMP contents in isolated vacuoles from 35S:ENT1 over-expresser mutants compared with WT plants. Finally, (v) a markedly increased release of labelled adenosine from preloaded liposomes containing vacuolar membrane protein from 35S:ENT1 over-expresser mutants compared with liposomes containing protein from WT or ENT1-RNAi vacuolar membranes was observed. As a result of increased nucleoside export from the vacuole, 35S:ENT1 mutants exhibit delayed vegetative growth on soil and increased activities of the salvage pathway enzyme adenosine kinase (AK).
The results presented here lead to the assumption that adenosine (and also other nucleosides, which could not be detected because of technical reasons), which appears as a breakdown product from RNA in the vacuole, is exported to the cytosol by ENT1 where it is metabolized with high affinity (Km 0.3–0.5 μM) by adenosine kinase (Moffatt et al., 2002). Considering the relative high vacuolar (73%) and low cytosolic (6.7%) volumes of plant cells (Winter et al., 1993) it becomes manifest that export of adenosine from the vacuole may lead to lowered total adenosine contents. Nevertheless, the cytosolic adenosine content may be increased in 35S:ENT1 mutants. It is known that adenosine kinase activity and thus the cytosolic adenosine contents are critical for the performance of methylation reactions through S-adenosylmethionine, because adenosine is a potent inhibitor thereof (Moffatt et al., 2002). This may be one explanation why these mutants show growth retardations, although further work on this issue has to be done in future. The measured increase of AK activity in 35S:ENT1 mutants can be interpreted as a compensatory reaction against increased cytosolic adenosine concentrations. A second compensatory reaction towards increased ENT1 expression was observed with a simultaneous increase in ENT7 expression in the vegetative tissues of corresponding mutants (Fig. 2B). Despite multiple approaches to determine the subcellular localization of ENT7 by use of GFP fusion constructs, we have failed so far. However, there are no indications for an organellar localization based on bioinformatic data, which favours the view of a localization of ENT7 in the secretory pathway. However, ENT7 residing in the plasma membrane or the vacuolar membrane is ideally suited to remove excess adenosine from the cytosol. This might explain the marked increase in ENT7 transcript in 35S:ENT1 mutants, although it has to be kept in mind that the absolute amount of ENT7 transcript is about 900 times lower, compared to ENT1.
Tissue-specific expression of ENT1
ENT1 expression was analysed with the help of promoter–GUS lines. ENT1 expression was found in distinct regions of hte roots of germinating seedlings and later on in development in the whole root cortex and the cotyledons. This observation is in line with microarray data showing a relatively uniform transcript pattern in root tissues with a slight increased accumulation in the root hair zone (Zimmermann et al., 2004). A supposed function of ENT1 in RNA turnover and hte export of breakdown products from the vacuole can be regarded as a housekeeping function of cells and thus fits to the quite broad expression pattern observed for this gene. ENT1 was expressed in nearly all plant organs but was especially marked in pollen. Quite a lot of transport proteins have been identified to be specifically expressed in pollen. Examples are NsAAP1, an amino acid permease, STP11, a monosaccharide transporter, GPT2, a plastidic glucose-6-phosphate transporter, and AAC4, a ER-localized ATP transporter (Lalanne et al., 1997; Niewiadomski et al., 2005; Schneidereit et al., 2005; Leroch et al., 2008). AtPTR5 is a dipeptide transporter also residing in pollen and affecting pollen germination and elongation (Komarova et al., 2008). However, in most cases, the exact role of the aforementioned proteins for pollen function is unknown. By contrast, ENT1 is clearly important for pollen germination. RNAi lines with reduced ENT1 show substantially decreased pollen germination rates. Under in vitro conditions, WT pollen germinate at a rate of 52–56%, which is in good accordance to other reports (Steinebrunner et al., 2003; Reichler et al., 2009) whereas RNAi pollen of all three lines analysed exhibit germination rates of only 38–41% (Fig. 5A). The reduced germination rate of ENT1-RNAi pollen was accompanied by reduced internal ATP contents as well as reduced external ATP levels (eATP). Assuming ENT1 resides in the vacuolar membrane of pollen, the export of nucleosides may be limited, leading to the observed higher overall and vacuolar levels of adenoaine and 2'3'-cAMP (Fig. 3A–C). In 35S:ENT over-expresser mutants, a higher capacity to salvage adenosine was observed. Therefore, it is likely that the reduced export of adenosine from ENT1-RNAi mutants leads to reduced salvage and thus to the measured reduced internal ATP levels. The importance of purine salvage for pollen metabolism has been demonstrated by knockout mutants lacking adenine phosphoribosyl transferase as these are male sterile (Moffatt and Sommerville, 1988). Taken together it can be assumed that both the salvage of purine nucleobases as well as salvage of purine nucleosides is critical for pollen metabolism. Furthermore, the function of external ATP for pollen germination has to be discussed in the context of ENT1 activity. It can be assumed that the level of internal ATP affects the level of eATP, although the export route for ATP is still under debate (Roux and Steinebrunner, 2007; Tanaka et al., 2010). External ATP and the activity of the enzyme which removes ATP, apyrase, markedly affect pollen germination (Steinebrunner et al., 2003). Thereby, ATP functions as a signal for so far unknown receptors. Increasing external concentrations of ATP inhibit pollen germination. Based on the model of Roux and Steinebrunner (2007), eATP affects growth in a concentration-dependent manner with an optimal concentration for growth and higher and lower eATP concentrations can negatively affect growth. Therefore, the reduced germination rate of ENT1-RNAi mutants may be a result of the observed lower eATP concentrations (Figs 7B, 8). The observed lowered uptake rates for adenosine in ENT1-RNAi pollen of line #42 may result from the down-regulation of other ENT transport proteins present at the pollen plasma membrane (an increased expression of ENT7 was observed for 35S:ENT1 over-expresser lines, Fig. 2A, but no alteration was observed in pollen from ENT1-RNAi line #42, data not shown). The observation that pollen from ENT1-RNAi lines almost exclusively could be isolated in the form of tetrads (Fig. 5B, C) was unexpected. A similar phenotype was observed in the Quartet1 mutant, lacking a pectin methylesterase (Francis et al., 2006). This enzyme activity is essential for pectin degradation and the release of single pollen from the quartet state during development. Whether cytosolic adenosine contents, known to affect the methylation capacity of cells and especially the level of methylesterified pectin in seed mucilage (Moffatt et al., 2002), also interact with the structure of pollen pectin thus influencing pollen release from the the quartet state remains to be analysed in future.
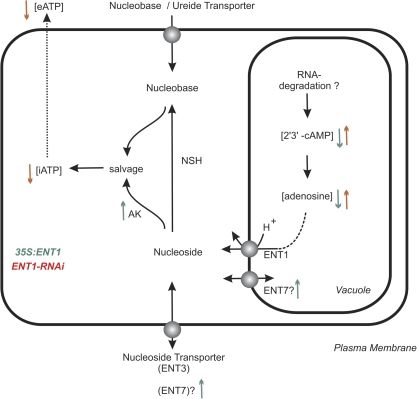
Fig. 8.
Model explaining the cellular alterations in ENT1 mutants. Arrows indicate up- or down-regulated enzyme activities (AK), transcript levels (ENT7) or metabolite concentrations. Colours indicate whether the measurements were obtained on 35S:ENT1 mutants relative to WT (green) or ENT1-RNAi mutants relative to WT (red). ENT7 appears at the plasmamembrane and the tonoplast as the subcellular localization has not been clarified. AK, adenosine kinase; NSH, nucleoside hydrolase; iATP, internal ATP; eATP, external ATP. (This figure is available in colour at JXB online.)
In sum, the following model is proposed: ENT1 releases adenosine and other nucleosides stemming from vacuolar RNA degradation (Fig. 8). These nucleosides can then be salvaged by nucleoside kinases (in the case of purine nucleosides by AK) or the concerted action of nucleoside hydrolase (NSH) and (adenine)-phosphoribosyltransferase (APRT). The salvage reactions affect internal and external ATP levels, whereas eATP regulates growth (Fig. 8). External ATP levels together with the effects of cytosolic adenosine negatively acting on S-adenosylmethionine-dependent methylation reactions provoke the observed phenotypes of ENT1 mutants as growth restrictions, reduced pollen germination, and the appearance of mature pollen as tetrads.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. PCR-primers used in this study.
Supplementary Fig. S1.Expression levels of ENTs in seedlings.
Supplementary Fig. S2. Light micrograph image of isolated vacuoles from Arabidopsis and immunodetection of ENT1 in these vacuolar preparations.
Acknowledgments
This work was financially funded by Deutsche Forschungsgemeinschaft (grant MO 1032/3-1 and STE A455/4-A) and the ‘Research Initiative Membrane Biology’ at the University of Kaiserslautern. HHK thanks the Alexander von Humboldt-Foundation and the Human Frontier Science Program for financial support. We gratefully acknowledge support of the work by Professor HE Neuhaus
Glossary
Abbreviations
- DW
dry weight
- ENT
equilibrative nucleoside transporter
- FW
fresh weight
- WT
wild type
References
- Abel S, Blume B, Glund K. Evidence for RNA-oligonucleotides in plant vacuoles isolated from cultured tomato cells. Plant Physiology. 1990;94:1163–1171. doi: 10.1104/pp.94.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Glund K. Localization of RNA-degrading enzyme activity within vacuoles of cultured tomato cells. Physiologia Plantarum. 1986;66:79–86. [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Molecular Biology. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Berthome R, Thomasset M, Maene M, Bourgeois N, Froger N, Budar F. Pur4 mutations are lethal to the male, but not the female, gametophyte and affect sporophyte development in Arabidopsis. Plant Physiology. 2008;147:650–660. doi: 10.1104/pp.108.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnstock G. Purinergic signalling: past, present and future. Brazilian Journal of Medical and Biological Research. 2009;42:3–8. doi: 10.1590/s0100-879x2008005000037. [DOI] [PubMed] [Google Scholar]
- Chen KL, Xu MX, Li GY, Liang H, Xia ZL, Liu X, Zhang JS, Zhang AM, Wang DW. Identification of ENT3 as the main transporter for uridine uptake in Arabidopsis roots. Cell Research. 2006;16:377–388. doi: 10.1038/sj.cr.7310049. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Demidchik V, Nichols C, Oliynyk M, Dark A, Glover BJ, Davies JM. Is ATP a signaling agent in plants? Plant Physiology. 2003;133:456–461. doi: 10.1104/pp.103.024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP. Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiology. 2006;142:1004–1013. doi: 10.1104/pp.106.085274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MR, Kim H, Kang SW, Kim JS, Jeong YM, Hwang HJ, Lee SY, Woo JC, Kim SG. Functional characterization of a gene encoding a dual domain for uridine kinase and uracil phosphoribosyltransferase in Arabidopsis thaliana. Plant Molecular Biology. 2007;63:465–477. doi: 10.1007/s11103-006-9101-3. [DOI] [PubMed] [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J. A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Moecular Cell Proteomics. 2007;6:394–412. doi: 10.1074/mcp.M600250-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeter CR, Tang W, Henaff E, Butterfield T, Roux SJ. Evidence of a novel cell signaling role for extracellular adenosine triphosphates and diphosphates in Arabidopsis. The Plant Cell. 2004;16:2652–2664. doi: 10.1105/tpc.104.023945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B, Flörchinger M, Kunz HH, Traub M, Wartenberg R, Jeblick W, Neuhaus HE, Möhlmann T. Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. The Plant Cell. 2009;21:876–891. doi: 10.1105/tpc.108.062612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B, Hoffmann C, Möhlmann T. Arabidopsis nucleoside hydrolases involved in intracellular and extracellular degradation of purines. The Plant Journal. 2011;65:703–711. doi: 10.1111/j.1365-313X.2010.04455.x. [DOI] [PubMed] [Google Scholar]
- Kamboj RK, Jackson JF. Pyrimidine nucleoside uptake by petunia pollen. Plant Physiology. 1985;79:801–805. doi: 10.1104/pp.79.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj RK, Jackson JF. Purine nucleoside transport in petunia pollen is an active, carrier-mediated system not sensitive to nitrobenzylthioinosine and not renewed during pollen tube growth. Plant Physiology. 1987;84:688–691. doi: 10.1104/pp.84.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Sivaguru M, Stacey G. Extracellular ATP in plants. Visualization, localization, and analysis of physiological significance in growth and signaling. Plant Physiology. 2006;142:984–992. doi: 10.1104/pp.106.085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova NY, Thor K, Gubler A, Meier S, Dietrich D, Weichert A, Suter GM, Tegeder M, Rentsch D. AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiology. 2008;148:856–869. doi: 10.1104/pp.108.123844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne E, Mathieu C, Roche O, Vedel F, De PR. Structure and specific expression of a Nicotiana sylvestris putative amino-acid transporter gene in mature and in vitro germinating pollen. Plant Molecular Biology. 1997;35:855–864. doi: 10.1023/a:1005812419151. [DOI] [PubMed] [Google Scholar]
- Leinhos V, Krauss GJ, Glund K. Evidence that a part of cellular uridine of a tomato (Lycopersicon esculentum) cell suspension culture is located in the vacuole. Plant Science. 1986;47:15–20. [Google Scholar]
- Leroch M, Kirchberger S, Haferkamp I, Wahl M, Neuhaus HE, Tjaden J. Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum. Journal of Biological Chemistry. 2005;280:17992–18000. doi: 10.1074/jbc.M412462200. [DOI] [PubMed] [Google Scholar]
- Leroch M, Neuhaus HE, Kirchberger S, Zimmermann S, Melzer M, Gerhold J, Tjaden J. Identification of a novel adenine nucleotide transporter in the endoplasmic reticulum of Arabidopsis. The Plant Cell. 2008;20:438–451. doi: 10.1105/tpc.107.057554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang D. Cloning and in vitro expression of the cDNA encoding a putative nucleoside transporter from Arabidopsis thaliana. Plant Science. 2000;157:23–32. doi: 10.1016/s0168-9452(00)00261-2. [DOI] [PubMed] [Google Scholar]
- Li G, Liu K, Baldwin SA, Wang D. Equilibrative nucleoside transporters of Arabidopsis thaliana: cDNA cloning, expression pattern and analysis of transport activities. Journal of Biological Chemistry. 2003;278:35732–35742. doi: 10.1074/jbc.M304768200. [DOI] [PubMed] [Google Scholar]
- Löffler A, Abel S, Jost W, Beintema JJ, Glund K. Phosphate-regulated induction of intracellular ribonucleases in cultured tomato (Lycopersicon esculentum) cells. Plant Physiology. 1992;98:1472–1478. doi: 10.1104/pp.98.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainguet SE, Gakiere B, Majira A, Pelletier S, Bringel F, Guerard F, Caboche M, Berthome R, Renou JP. Uracil salvage is necessary for early Arabidopsis development. The Plant Journal. 2009;60:280–291. doi: 10.1111/j.1365-313X.2009.03963.x. [DOI] [PubMed] [Google Scholar]
- Mansfield TA, Schultes NP, Mourad GS. AtAzg1 and AtAzg2 comprise a novel family of purine transporters in Arabidopsis. FEBS Letters. 2009;583:481–486. doi: 10.1016/j.febslet.2008.12.048. [DOI] [PubMed] [Google Scholar]
- Moffat B, Sommerville C. Positive selection for male-sterile mutants of Arabidopsis lacking adenine phophoribosyl transferase activity. Plant Physiology. 1988;86:1150–1154. doi: 10.1104/pp.86.4.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, Stevens YY, Allen MS, Snider JD, Pereira LA, Todorova MI, Summers PS, Weretilnyk EA, Martin-McCaffrey L, Wagner C. Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiology. 2002;128:812–821. doi: 10.1104/pp.010880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhlmann T, Batz O, Maaß U, Neuhaus HE. Analysis of carbohydrate transport across the envelope of isolated cauliflower-bud amyloplasts. Biochemical Journal. 1995;307:521–526. doi: 10.1042/bj3070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhlmann T, Mezher Z, Schwerdtfeger G, Neuhaus HE. Characterisation of a concentrative type of adenosine transporter from Arabidopsis thaliana (ENT1, At) FEBS Letters. 2001;509:370–374. doi: 10.1016/s0014-5793(01)03195-7. [DOI] [PubMed] [Google Scholar]
- Möhlmann T, Bernard C, Hach S, Neuhaus HE. Nucleoside transport and associated metabolism. Plant Biology. 2010;12:26–34. doi: 10.1111/j.1438-8677.2010.00351.x. [DOI] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge UI, Schneider A. The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. The Plant Cell. 2005;17:760–775. doi: 10.1105/tpc.104.029124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichler SA, Torres J, Rivera AL, Cintolesi VA, Clark G, Roux SJ. Intersection of two signalling pathways: extracellular nucleotides regulate pollen germination and pollen tube growth via nitric oxide. Journal of Experimental Botany. 2009;60:2129–2138. doi: 10.1093/jxb/erp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser J, Linka N, Lemke L, Jeblick W, Neuhaus HE. Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiology. 2004;136:3524–3536. doi: 10.1104/pp.104.049502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Zouhar J, Carter C, Raikhel N. Isolation of intact vacuoles from Arabidopsis rosette leaf-derived protoplasts. Nature Protocols. 2007;2:259–262. doi: 10.1038/nprot.2007.26. [DOI] [PubMed] [Google Scholar]
- Roux SJ, Steinebrunner I. Extracellular ATP: an unexpected role as a signaler in plants. Trends in Plant Science. 2007;12:522–527. doi: 10.1016/j.tplants.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Su YH, Kunze R, Warner S, Hewitt M, Slocum RD, Ludewig U, Frommer WB, Desimone M. UPS1 and UPS2 from Arabidopsis mediate high affinity transport of uracil and 5-fluorouracil. Journal of Biological Chemistry. 2004;279:44817–44824. doi: 10.1074/jbc.M405433200. [DOI] [PubMed] [Google Scholar]
- Schneidereit A, Scholz-Starke J, Sauer N, Büttner M. AtSTP11, a pollen tube-specific monosaccharide transporter in Arabidopsis. Planta. 2005;221:48–55. doi: 10.1007/s00425-004-1420-5. [DOI] [PubMed] [Google Scholar]
- Steinebrunner I, Wu J, Sun Y, Corbett A, Roux SJ. Disruption of apyrases inhibits pollen germination in Arabidopsis. Plant Physiology. 2003;131:1638–1647. doi: 10.1104/pp.102.014308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Gilroy S, Jones AM, Stacey G. Extracellular ATP signaling in plants. Trends in Cell Biology. 2010;20:601–608. doi: 10.1016/j.tcb.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub M, Flörchinger M, Piecuch J, Kunz H-H, Weise-Steinmetz A, Deitmer JW, Neuhaus HE, Möhlmann T. The fluorouridine insensitive 1 (fur1) mutant is defective in equilibrative nucleoside transporter 3 (ENT3), and thus represents an important pyrimidine nucleoside uptake. The Plant Journal. 2007;49:855–864. doi: 10.1111/j.1365-313X.2006.02998.x. [DOI] [PubMed] [Google Scholar]
- Trentmann O, Horn M, van Scheltinga AC, Neuhaus HE, Haferkamp I. Enlightening energy parasitism by analysis of an ATP/ADP transporter from chlamydiae. PLoS.Biology. 2007;5:e231. doi: 10.1371/journal.pbio.0050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers MF, Yao SYM, Baldwin SA, Young JD, Cass CE. Nucleoside transporter proteins of Saccharomyces cerevisiae. Journal of Biological Chemistry. 2000;275:25931–25939. doi: 10.1074/jbc.M000239200. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA. Construct design for efficient, effective and high-throughput gene silencing in plants. The Plant Journal. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Winter H, Robinson DG, Heldt HW. Subcellular volumes and metabolite concentrations in barley leaves. Planta. 1993;191:180–190. [Google Scholar]
- Wolf C, Hennig M, Romanovicz D, Steinebrunner I. Developmental defects and seedling lethality in apyrase AtAPY1 and AtAPY2 double knockout mutants. Plant Molecular Biology. 2007;64:657–672. doi: 10.1007/s11103-007-9184-5. [DOI] [PubMed] [Google Scholar]
- Wormit A, Traub M, Flörchinger M, Neuhaus HE, Möhlmann T. Characterization of three novel members of the Arabidopsis thaliana equilibrative nucleoside transporter (ENT) family. Biochemical Journal. 2004;383:19–26. doi: 10.1042/BJ20040389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Steinebrunner I, Sun Y, Butterfield T, Torres J, Arnold D, Gonzalez A, Jacob F, Reichler S, Roux SJ. Apyrases (nucleoside triphosphate-diphosphohydrolases) play a key role in growth control in Arabidopsis. Plant Physiology. 2007;144:961–975. doi: 10.1104/pp.107.097568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Osakabe Y, Mizoi J, Nakashima K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis thaliana abiotic stress-inducible facilitated diffusion transporter for monosaccharides. Journal of Biological Chemistry. 2010;285:1138–1146. doi: 10.1074/jbc.M109.054288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Henning L, Gruissem W. Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R. Pyrimidine and purine biosynthesis and degradation in plants. Annual Review of Plant Biology. 2006;57:805–836. doi: 10.1146/annurev.arplant.57.032905.105421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.