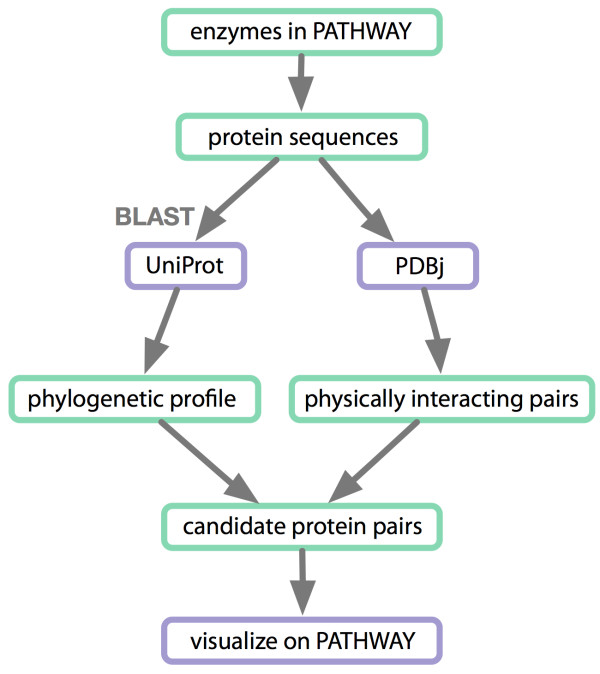
Figure 4.
Workflow to analyze protein interactions among enzymes in a KEGG pathway. First, protein sequences are retrieved for each enzyme in a KEGG pathway. The sequences are then BLAST searched against UniProt and a phylogenetic profile is constructed of the results. Then, for each species in the phylogenetic profile, BLAST searches are run against PDB. Pairs of protein sequences (of the same species) that have homologs in the same PDB entry are inferred to be in physical contact and hence predicted to be interacting. Conserved and interacting proteins are then visualized on the pathway map, an example of which is shown in Figure 5.