Figure 3.
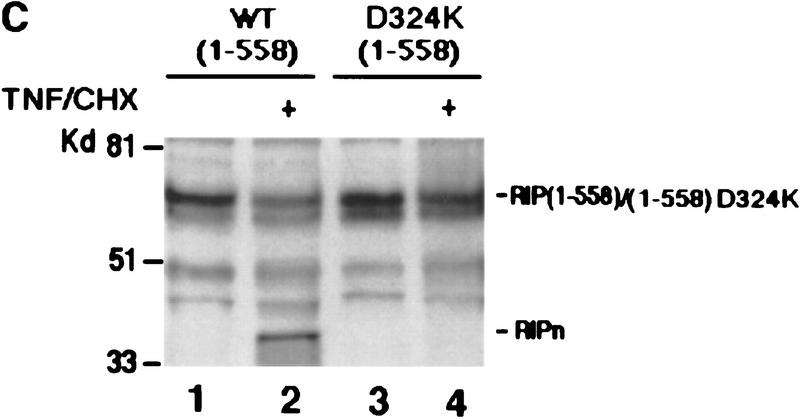
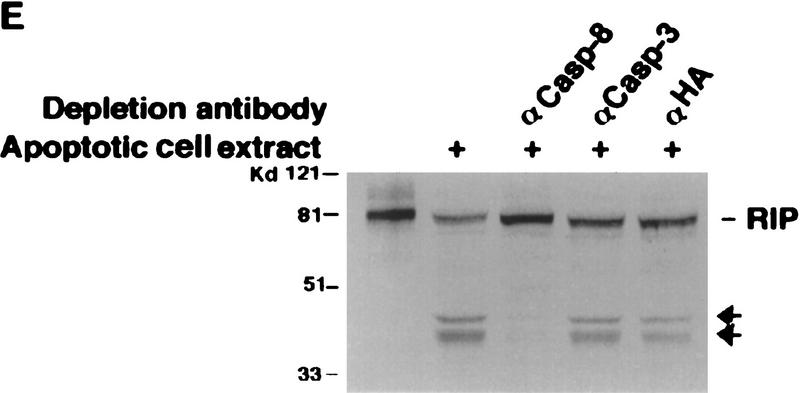
Mapping of the caspase cleavage site in RIP. (A) Schematic illustration of substitution mutants of RIP constructs. The kinase and death domains of RIP are shown as shaded and solid bars, respectively. The amino acid substitutions of each mutant are indicated. (B) 35S-Labeled RIP proteins were incubated with apoptotic HeLa cell extract (lanes 3,6,9,12) or normal HeLa cell extract (lanes 2,5,8,11) at 30°C for 3 hr, resolved on SDS-PAGE and visualized by autoradiograph. Nontreated RIP proteins were loaded as controls (lanes 1,4,7,10). (Lanes 1–3) Wild-type RIP; (lanes 4–6) RIP(D324K); (lanes 7–9) RIP (D248A, D250A, D252P); (lanes 10–12) RIP(D300V). The resultant cleaving products are indicated with arrows. (C) HeLa cells were transfected with Myc-tagged RIP(1–558) (lanes 1,2) or RIP(1–558, D324K) (lanes 3,4). Cells were treated with TNF + CHX (lanes 2,4) or remained untreated (lanes 1,3). Myc-tagged RIP proteins were detected by Western blot analysis with anti-Myc antibody. The position of Myc-tagged RIP (1–558)/ RIP (1–558, D324K) is indicated. RIPn indicates the resultant Myc-tagged amino-terminal portion of RIP (1–324 amino acids). (D) 35S-Labeled RIP proteins were incubated with Caspase-3 (lanes 2,6), Caspase-8 (lanes 3,7), or apoptotic HeLa cell extract at 37°C for 2 hr, resolved on SDS-PAGE and visualized by autoradiograph. Nontreated RIP proteins were loaded as controls (lanes 1,5). The resultant cleavage products are indicated with arrowheads. (E) Fifty microliters of apoptotic cell extract was incubated with the indicated antibodies bound on protein A resin at 4°C for 12 hr. The resin was removed by centrifugation and the supernatants were applied in cleavage assays as described in B. The positions of the molecular mass markers are indicated in kD at left of panels.