Figure 4.
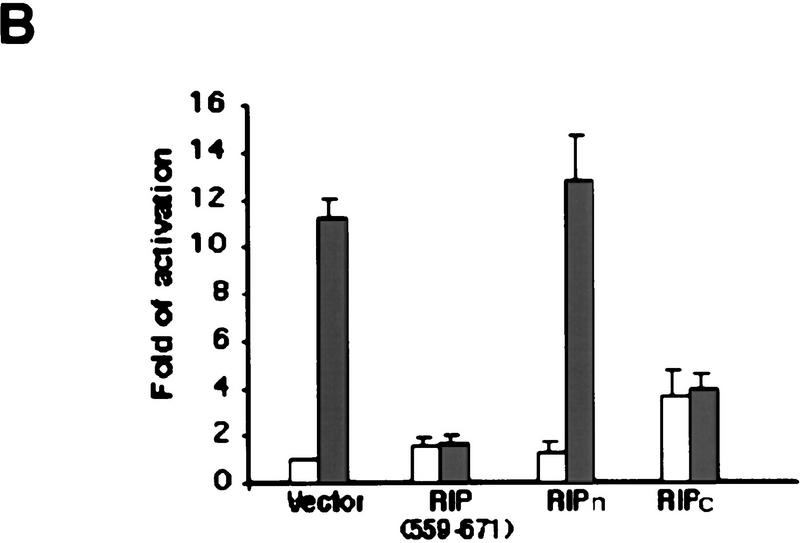
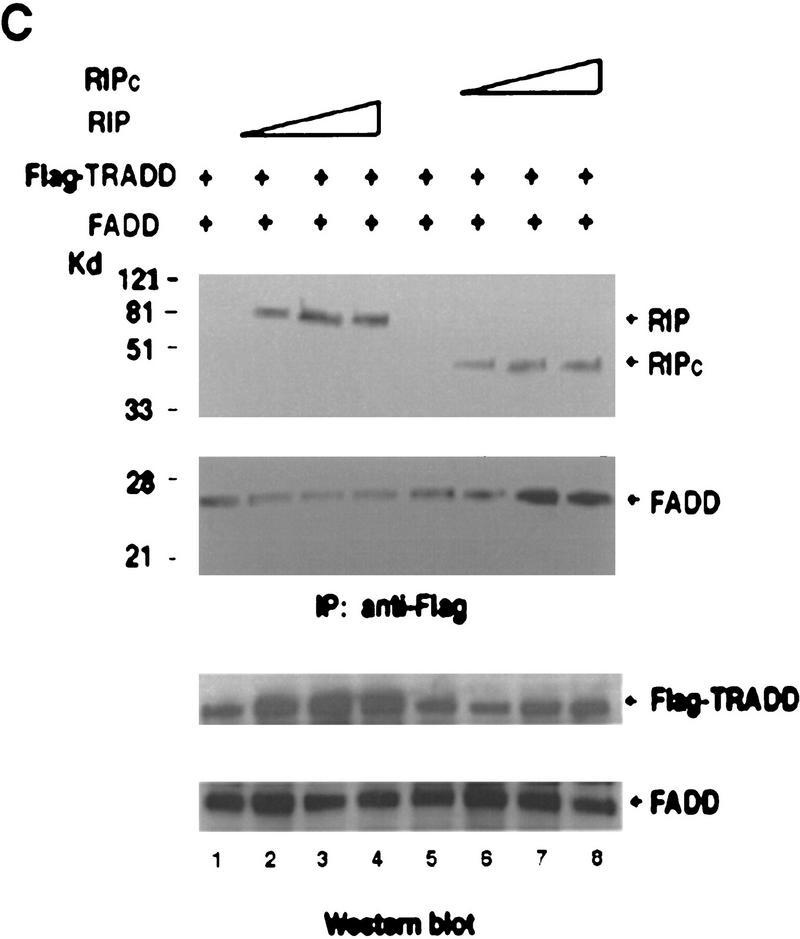
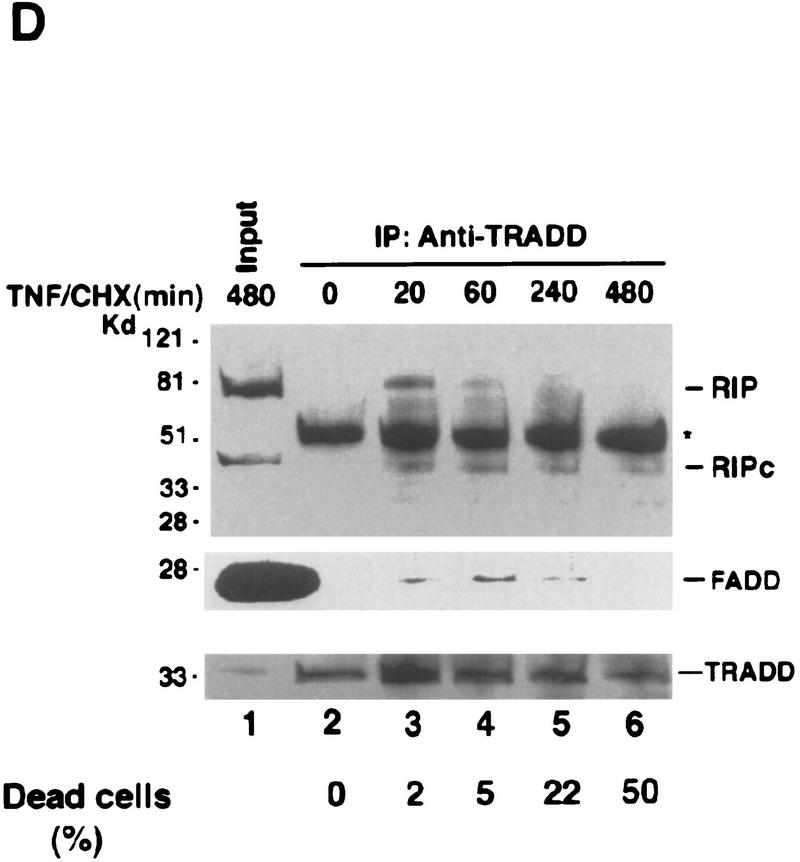
Effects of RIP cleavage on NF-κB activation and TRADD/FADD interaction. (A) Cells were co-transfected with p2xNF-κB–Luc, pRSV–lacZ and different RIP constructs as indicated or an empty vector. In the experiments described in the right panel, the expression vector of CrmA was also included. Twenty-four hours post-transfection, the cells were collected. Luciferase assay was conducted and normalized as described (Liu et al. 1996). The results were the average of three duplicated experiments. (B) Cells were cotransfected with p2xNF-κB–Luc, CrmA and different RIP constructs. Twenty-four hours post-transfection, half of the cells were treated with TNF (15 ng/ml) for 10 hr (solid bars). (Open bars) Nontreated cells. Luciferase activities were detected and normalized. The results shown were the average of three duplicated experiments. (C) HEK293 cells were cotransfected with FLAG–TRADD (2.5 μg), FADD (2.5 μg), CrmA (1 μg), along with increasing amounts of Xpress-tagged RIP (1.5, 3, 4.5 μg in lanes 2, 3, and 4, respectively) or RIPc (1.5, 3, 4.5 μg in lanes 6, 7, and 8, respectively). The total amount of DNA in each transfection was adjusted to 10 μg with the empty vector. Cells were collected at 24 hr after transfection. Expressions of Flag–TRADD and FADD were determined by Western blot (bottom). Immunoprecipitation experiments were performed with anti-Flag (M2) antibody, and coprecipitated FADD and Xpress-tagged RIP proteins were detected by Western blotting with anti-FADD and anti-Xpress, respectively (top). (D). HeLa cells (2 × 107) were treated with TNF (15 ng/ml) + CHX (10 μg/ml) for the indicated time periods, immunoprecipitation experiments were performed with anti-TRADD antibody, and coprecipitated RIP, FADD, and TRADD proteins were detected by Western blot. (Lane 1) One percent of the cell extract from the 480-min sample as an input control. The blot of FADD was exposed for a longer time to visualize the precipitated FADD protein. Cell death was determined by trypan blue exclusion staining. The percentages of dead cells are shown at the bottom of the panel. The positions of the molecular mass markers are indicated in kD on the left of C and D. (*IgG heavy chain).