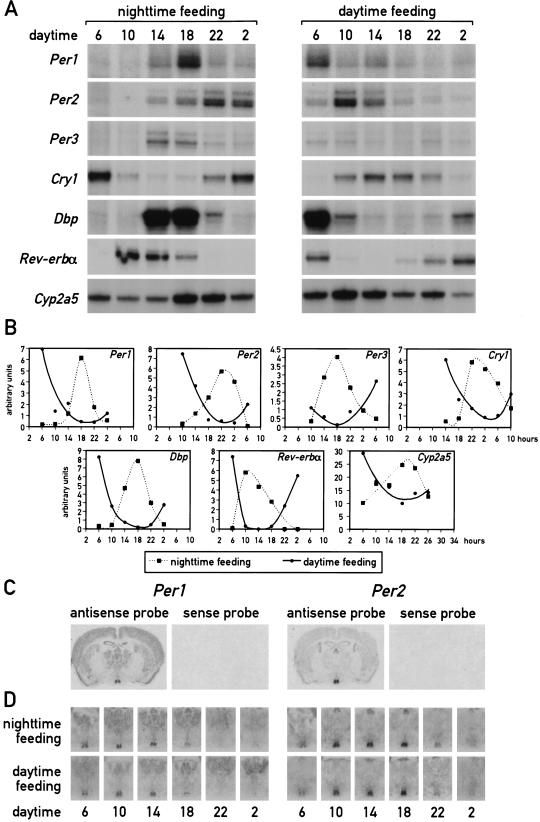
Figure 1.
Daytime feeding changes the phase of circadian gene expression in the liver but not in the suprachiasmatic nucleus (SCN). Mice, kept under a light-dark regimen (lights on 6 a.m., lights off 6 p.m.), were fed for eight consecutive days exclusively during the light phase (6 a.m. to 6 p.m.) or during the dark phase (6 p.m. to 6 a.m.). During the ninth day, animals were killed at 4-h intervals to prepare whole-cell RNA from liver and tissue sections from the brain. (A) Circadian accumulation of clock and clock-controlled genes in the liver. The levels of transcripts from the genes indicated to the left of the panels were determined by ribonuclease protection assays (Cyp2a5 stands for coumarin hydroxylase). Tbp mRNA or β-actin mRNA were included as standards for transcripts whose expression is constant throughout the day (data not shown). (B) The mRNA signals obtained in the ribonuclease protection assays shown in panel A were measured by scanning the autoradiography and normalized to the signals generated by β-actin mRNA (for Cyp2a5) or Tbp mRNA (for all other transcripts). (C) Serial coronal brain sections taken above the optical chiasma were hybridized in situ to 35S-labeled Per1 and Per2 antisense and sense RNA strands. Note that no detectable hybridization signals were obtained with the sense probe. (D) Circadian accumulation of Per1 and Per2 mRNA in the SCN of mice after 8 d of restricted feeding. Coronal sections from brains harvested at 4-h intervals around the clock were hybridized in situ to Per1 and Per2 antisense RNA probes as described in panel C. Note that the phases of rhythmic Per1 and Per2 expression are not affected by restricted feeding.