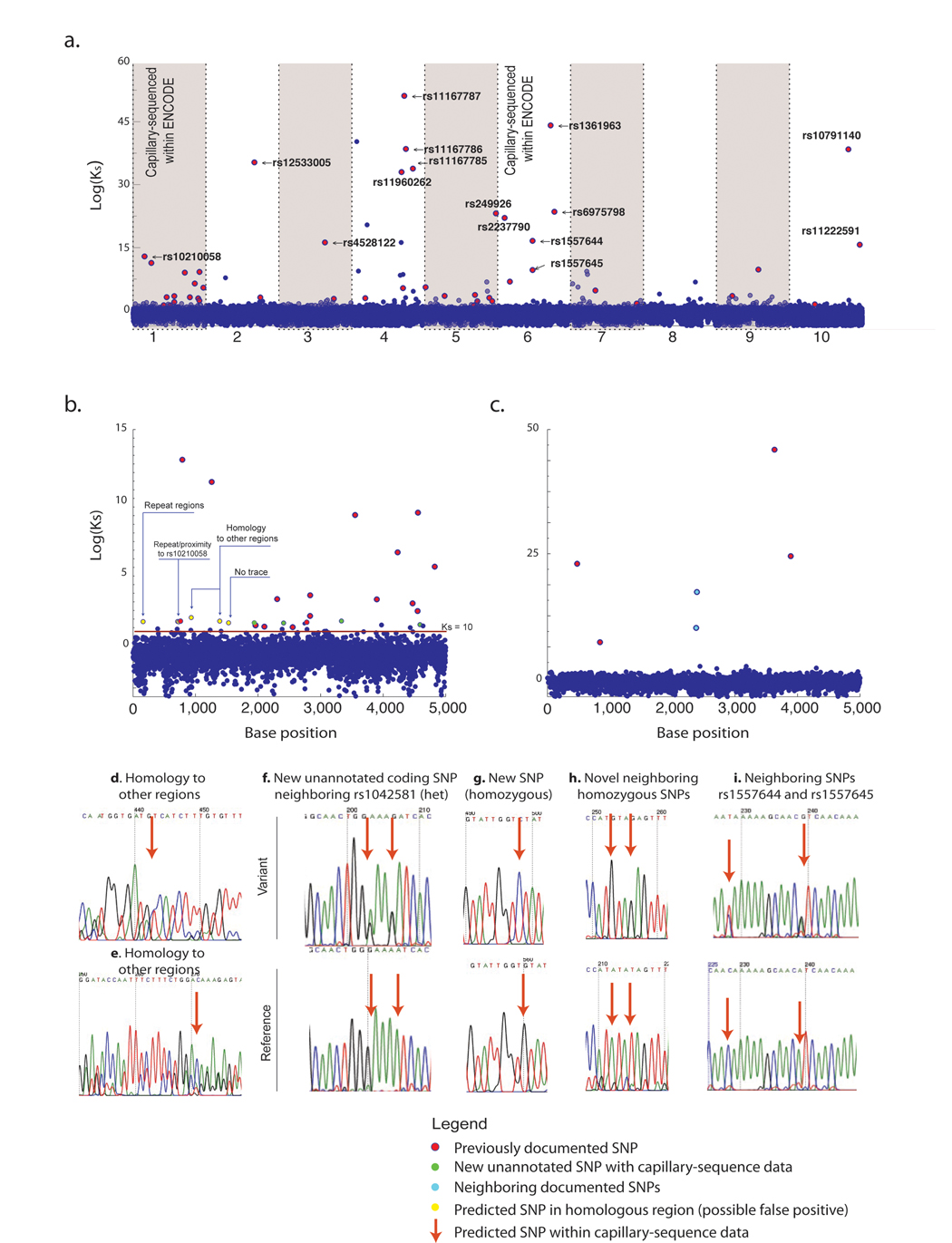
Figure 4. Discovery of variant bases by simultaneous analysis of all individuals.
(a.) The Bayes-factor for polymorphism discovery(Ks) is plotted for each of the10 sequenced 5kb amplicons from Library A. Exact positions matching known polymorphisms are colored as red spheres and the dbSNP identifier is provided for the most significant SNPs. Black bars at top indicate locations of documented SNPs. A magnified view of amplicon 1 (b.) and amplicon 6 (c.) is provided to compare variants predicted by indexed-multiplexed sequencing to previous deep capillary sequencing results for the same individuals as part of the ENCODE project. (d–e.) Examples of false-positives arising from sequence homology to elsewhere in the genome. (f–i.) Examples of sequence traces validating the discovery of novel SNPs not previously annotated in ENCODE capillary sequencing traces. Similar analysis was conducted on Library B (shown in the supplementary figure 1).