Figure 7.
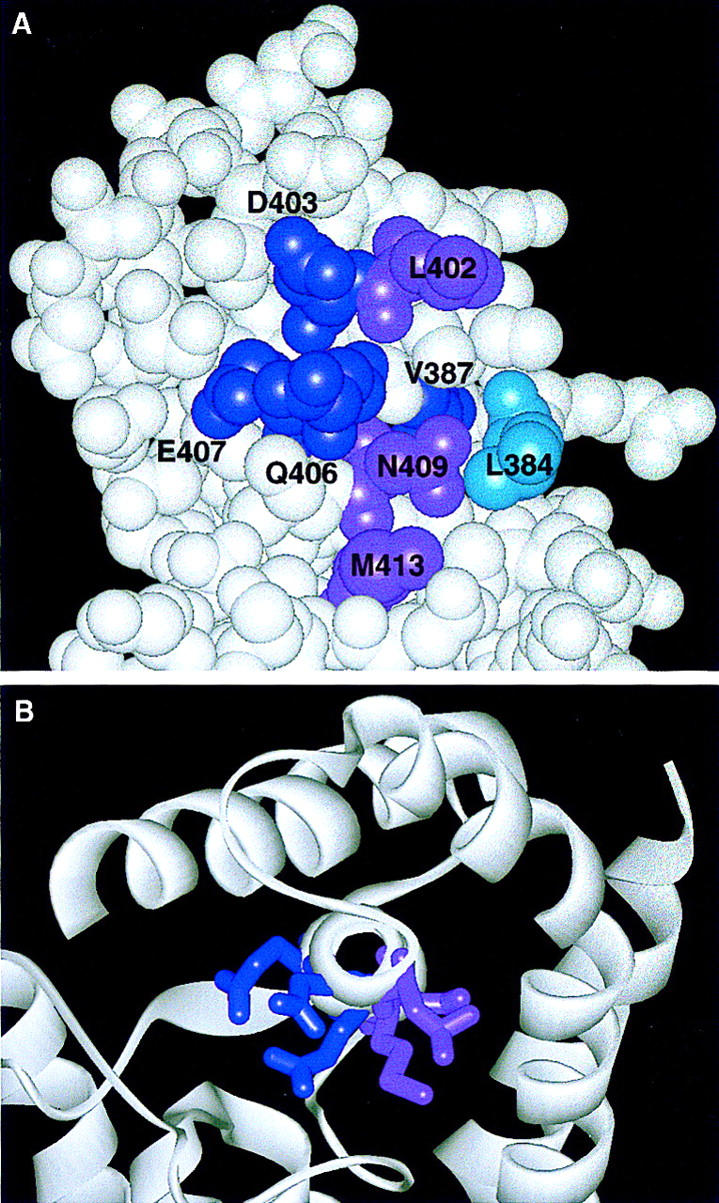
Location on the crystal structure of the σ70 residues implicated in core binding. (A) A space-filling model indicates residues from regions 2.1 and 2.2 implicated in core binding. Residues in dark purple were identified by genetic selection for relief of toxicity (V387, D403, and E407) or by a comparable σ32 selection (Q406); those in light purple were identified because of their defect in Q-mediated antitermination (L402, N409, and M413); the residue in cyan was proposed from the crystal structure (L384) (Malhotra et al. 1996). (B) A view of the σ70 crystal structure looking down or through helix 13 (region 2.2) showing that the region 2.2 mutants from the two different selections (indicated by light and dark purple) are localized on two different faces of the helix.
