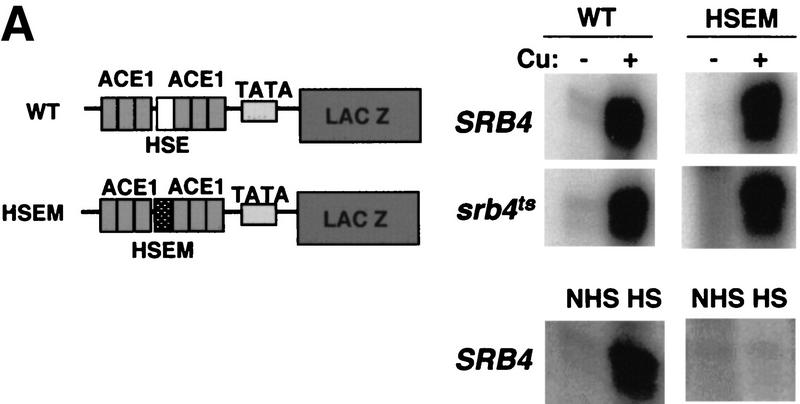
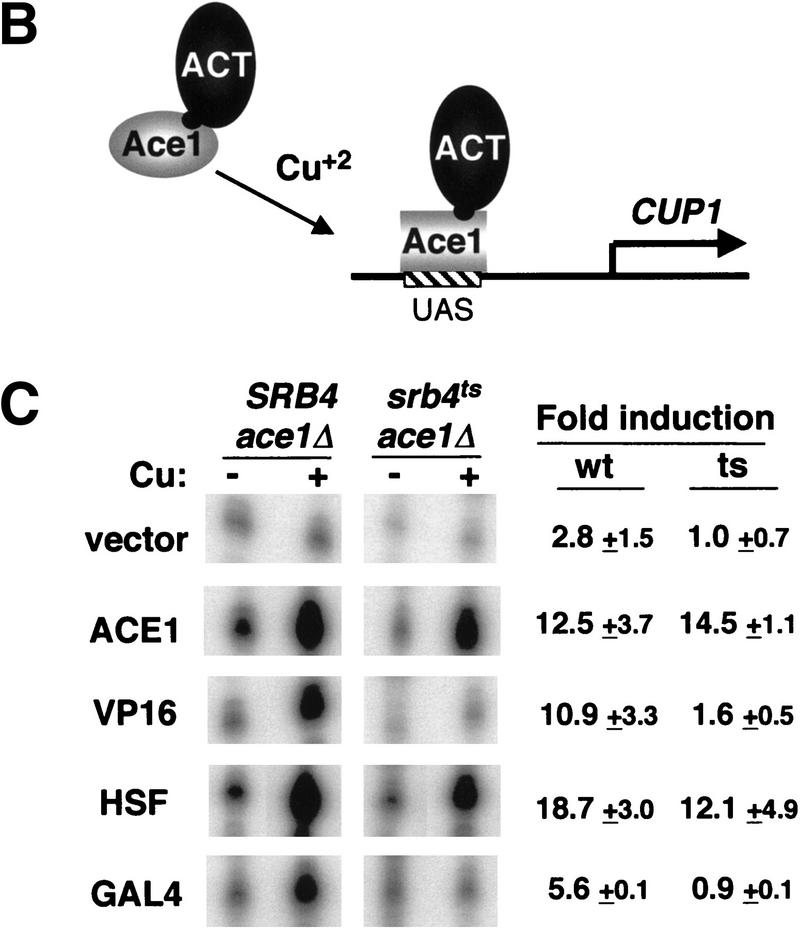
Figure 1.
Srb4 independence of the native CUP1 gene is activation domain specific. (A) S1 nuclease assays showing the heat shock element (HSE) is dispensable and Ace1-binding sites in the UAS of the CUP1 gene are sufficient for the Srb4-independent transcription. (Left) Reporter constructs used in the experiment. The binding sites for Ace1 (ACE1) and a single heat shock element (HSE), and TATA box (TATA) are shown. The HSE with a point mutation that destroys Hsf binding is designated as HSEM. (Top, right) The 30-min copper induction of wild-type (WT) and HSE mutant CUP1 promoter (HSEM) in SRB4 wild-type or srb4ts mutant. SRB4wt and srb4ts cells were grown until OD = 0.3–0.5 at 25°C, then cells were moved to a 37°C water bath and incubated for 1.5 hr; 8-ml cells were harvested (−Cu sample). Then, 1 mm CuSO4 was added and cells were incubated for 30 min at 37°C and again, 8-ml cultures were harvested (+Cu sample). (Bottom, right) The heat inducibility of each construct. Cells were grown to mid-log phase at 25°C and an aliquot was taken (NHS, non heat shock) or cells were transferred to a 39°C water bath for 15 min (HS, heat shock). RNAs were isolated and assayed by S1 nuclease protection. (B) Design of the experiment in part C. (C) Transcriptional activation of the CUP1 gene by Ace1-hybrid activators. Isogenic DLY981 (SRB4ace1Δ) and DLY982 (srb4tsace1Δ) strains are transformed with the following plasmids: pRS316 (vector), p316:ACE1 (ACE1), p316:ACVP (VP16), pAC–HSF (HSF), and pAC–GAL4 (GAL4). Cell growth and copper induction was done as in Fig. 1A. Fold induction of the endogenous CUP1 gene in wild-type and temperature-sensitive mutants are shown next to the S1 assay raw data. Standard deviation is shown for each value. Each value is an average of at least three experiments except for GAL4 (two).