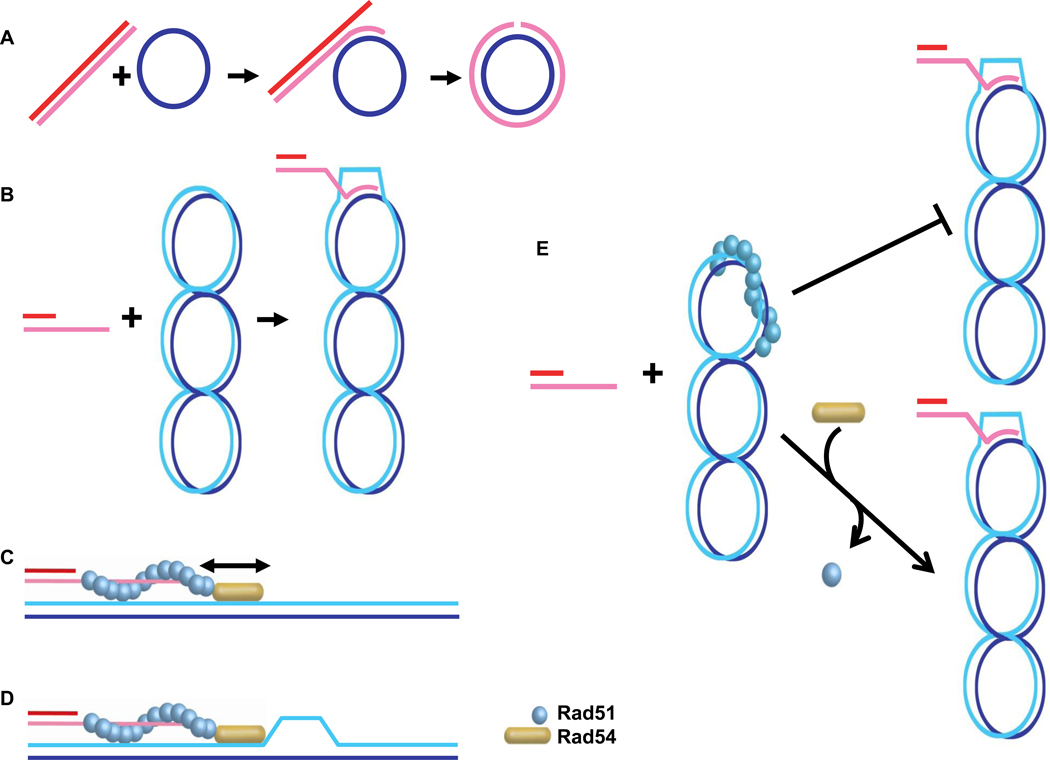
Figure 5. Homology search and DNA strand invasion.
A. Scheme of the three-stranded DNA strand exchange reaction between linear duplex DNA and circular single-stranded DNA. B. Scheme of the D-loop reaction between supercoiled, circular duplex DNA and a single-stranded DNA oligonucleotide, which is depicted with a heterologous duplex DNA tail. C. The dsDNA sliding model envisions that the Rad54 motor protein slides duplex DNA along the Rad51 presynaptic filament to enhance homology search. D. The dsDNA strand-opening model suggests that Rad54 translocation on the duplex target DNA results in strand opening that facilitates the conversion to stable joint molecules. In both models, Rad54 protein needs to transition from an ATPase inactive form when associated with the Rad51-ssDNA filament to an active ATPase on duplex DNA, a process which is not depicted in the figure. E. The Rad51-dsDNA complex dissociation model maintains that in the in vitro reaction some Rad51 will bind to the duplex DNA, which inhibits D-loop formation, unless Rad51 is dissociated from duplex DNA by Rad54 and entirely redistributed to bind to the ssDNA.