Figure 1.
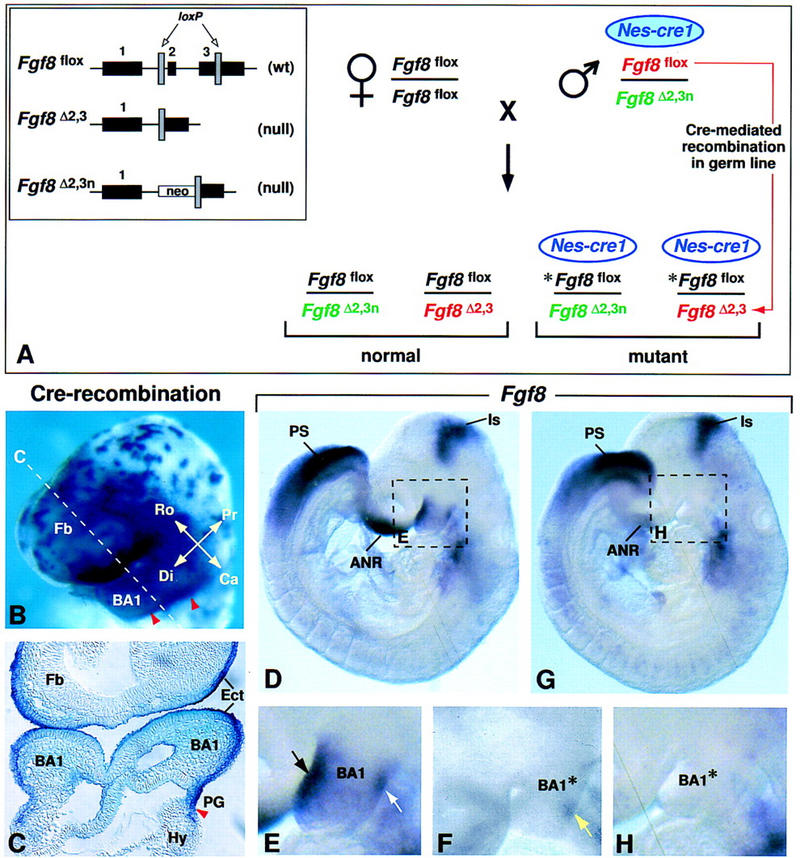
Inactivation of the Fgf8 gene in BA1 ectoderm. (A) Breeding scheme used to generate 50% mutant offspring. The three different Fgf8 alleles present in this cross are illustrated in the box on the left. The Fgf8flox allele has wild-type (wt) activity and can be converted to a null allele (Fgf8Δ2,3) by Cre-mediated recombination (Meyers et al. 1998). The Nes–cre1 gene used in this cross is imprinted, and is active in somatic tissues as described (see text) only when it is paternally inherited. However, it is active in the germ line irrespective of whether it is paternally or maternally inherited (A. Trumpp, B. Bates, R. Jaenisch, G.R. Martin, and J.M. Bishop, unpubl.). In the cross performed, the parental male has a normal phenotype because he carries a maternally inherited Nes–cre1 transgene (blue filled oval), which has little activity in somatic cells. When this transgene is transmitted to his offspring (unfilled oval), it is active in somatic tissues. In addition, this male transmits two different Fgf8 null alleles: Fgf8Δ2,3n (shown in green) and Fgf8Δ2,3 (shown in red), which is generated by Cre-mediated recombination in his germ line of the Fgf8flox allele he carries (also shown in red). Thus, all offspring carry a paternally inherited Fgf8 null allele, as well as a maternally inherited Fgf8flox allele (shown in black). In the 50% of offspring that inherit Nes–cre1, the floxed allele (*Fgf8flox) undergoes Cre-mediated recombination, resulting in tissue-specific inactivation of Fgf8. (B,C) Assay for Cre-mediated recombination using the Z/AP reporter gene (Lobe et al. 1999). Cells in which the Nes–cre1 gene was active produce human alkaline phosphatase (hAP). (B) Nes–cre1;Z/AP embryo at E9.0 (16 somites) stained for hAP activity in whole mount. The dashed line indicates the level of the section shown in C. The caudal limit of recombination (marked by red arrowheads) is located in the first pharyngeal groove. Note that recombination occurs in BA1 ectoderm. (D–H) Fgf8 expression as detected by whole mount RNA in situ hybridization in D and E control and F to H Fgf8;Nes–cre mutant embryos at E9.0 (14 somites). The dashed boxes in D and G indicate the regions shown at higher magnification in E and H, respectively. In the mutant embryo, the levels of Fgf8 RNA in most expression domains are similar to those in the normal embryo (D). However, they appear to be reduced in the front of the face/anterior neural ridge, and, significantly, no Fgf8 RNA is detected in BA1. Black and white arrows in E point to the normal Fgf8 expression domains on the rostral and caudal side of BA1, respectively. (F) Fgf8 RNA is detected on the caudal side of BA1 (yellow arrow) in a mutant embryo at a slightly earlier stage. (ANR) Anterior neural ridge; (BA1) first branchial arch; (BA1*) mutant BA1; (Ca) caudal; (Di) distal; (Ect) ectoderm; (Fb) forebrain; (Hy) hyoid arch (second branchial arch); (Is) isthmic constriction; (PG) pharyngeal groove; (Pr) proximal; (PS) primitive streak; (Ro) rostral.
