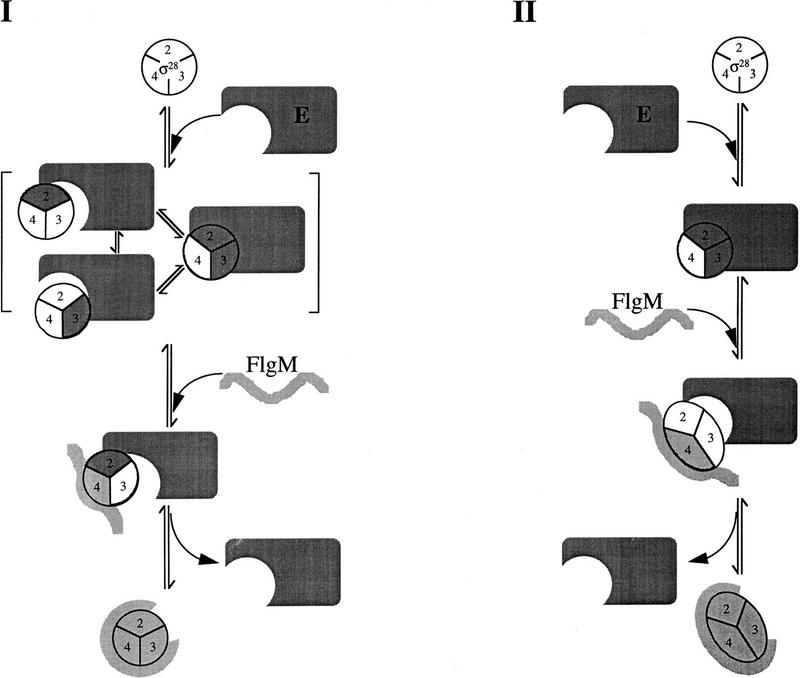
Figure 6.
Possible mechanisms for holoenzyme destabilization by FlgM. Conserved σ regions 2, 3, and 4 are numbered. σ28 regions interacting with core RNAP (E) are shaded dark gray; σ28 regions interacting with FlgM are shaded light gray. Free FlgM is depicted as an unstructured molecule; σ28-bound FlgM is depicted in a constrained conformation. For simplicity, only the carboxy-terminal σ28-binding domain of FlgM is shown participating in the interaction with σ28. (I) Competitive displacement model. Eσ28 breathing alternately exposes σ regions 2 and 3. FlgM partially bound to σ28 can compete with E for rebinding to regions 2 and 3 to effect dissociation. For simplicity, FlgM is only shown interacting with one of the two partially bound forms of σ28. (II) Allosteric displacement model. Binding of FlgM to exposed region 4 stabilizes a σ28 conformation that is incompatible with E, resulting in the release of the σ28–FlgM complex.