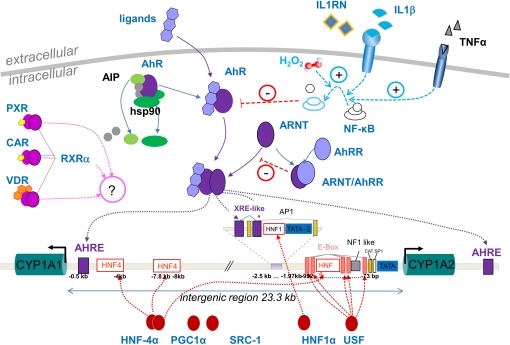
Figure 1.
Overview of known pathways for transcriptional regulation at the CYP1A2 promoter. The scheme shows the 23.3-kb intergenic region between the head-to-head oriented CYP1A1 and 1A2 genes on chromosome 15q24.1. Binding elements located within the common promoter region may thus be shared for transcriptional regulation of both genes. Pathways for constitutive and tissue specific expression are shown by red symbols at the bottom and include the liver-enriched transcription factors HNF4α and HNF1α and their coactivators PGC1α and SRC-1, as well as basic transcriptional activators SP-1 and USF-1 (Quattrochi et al., 1994; Chung and Bresnick, 1997; Narvaez et al., 2005; Martínez-Jiménez et al., 2006). Top-middle: Genes selected for the AhR-pathway include the ligand-binding receptor AhR, the AhR nuclear translocator ARNT, and the AhR regulator AhRR. The activated ligand bound form of AhR is complexed with two heat-shock proteins hsp90 and AIP, and co-chaperone p23, which help to correctly fold and stabilize the AhR and prevent inappropriate trafficking to the nucleus (Petrulis et al., 2000). Upon heterodimerization with activated AhR, the ARNT/AhR complex translocates to the nucleus to activate transcription from AHRE- and XRE-motifs (Okino et al., 2007). AhRR competes with AhR for ARNT binding, resulting in inhibition of AhR-mediated signal transduction. Top-right: Under pathophysiological conditions of inflammation, inflammatory cytokines like IL1ß and TNFα or chronic oxidative stress (H2O2) activate NFκB which in turn inhibits AhR activity thus leading to reduced CYP1A expression. This pathway can be blocked by competing antagonists like IL1RN (Tian et al., 1999; Vrzal et al., 2004; Zhou et al., 2008). Top-left: Exploratory pathways involving possible crosstalk to other known xenobiotic transcriptional regulation networks are indicated by PXR/RXRα, CAR/ RXRα, VDR/ RXRα heterodimers. Proteins designated in black were not included in this study.