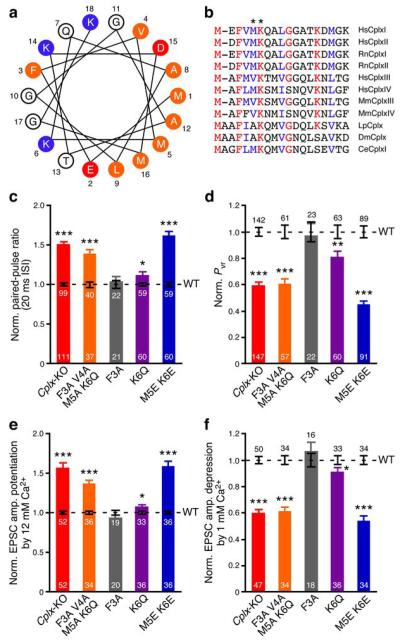
Figure 2.
Identification of Met5 and Lys6 as crucial residues for CplxI N-terminal function. (a) Helical wheel model of rat CplxI N terminus. Residues are numbered and labeled as orange (hydrophobic), red (negatively charged), and blue (positively charged). (b) N terminus amino acid sequence alignment of Complexin paralogs and orthologs. Identical residues are marked as red and highly conserved residues are marked as blue. Hs, Homo sapiens; Rn, Rattus norvegicus; Mm, Mus musculus; Lp, Loligo pealeii; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans. Asterisks mark residues Met5 and Lys6 in RnCplxI. (c–f) Summary data of evoked release from Cplx-KO neurons and Cplx-KO neurons rescued by WT or mutant CplxI. Bar graphs show paired-pulse ratio (c), Pvr (d), EPSC amplitude potentiation by elevating Ca2+ concentration (e), and EPSC amplitude depression by lowering Ca2+ concentration (f). Data are normalized to the mean values of the corresponding CplxI WT rescue (black dashed lines and error bars). Data are expressed as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.0001 compared to the corresponding CplxI WT rescue. The numbers of neurons analyzed are indicated on the bars.