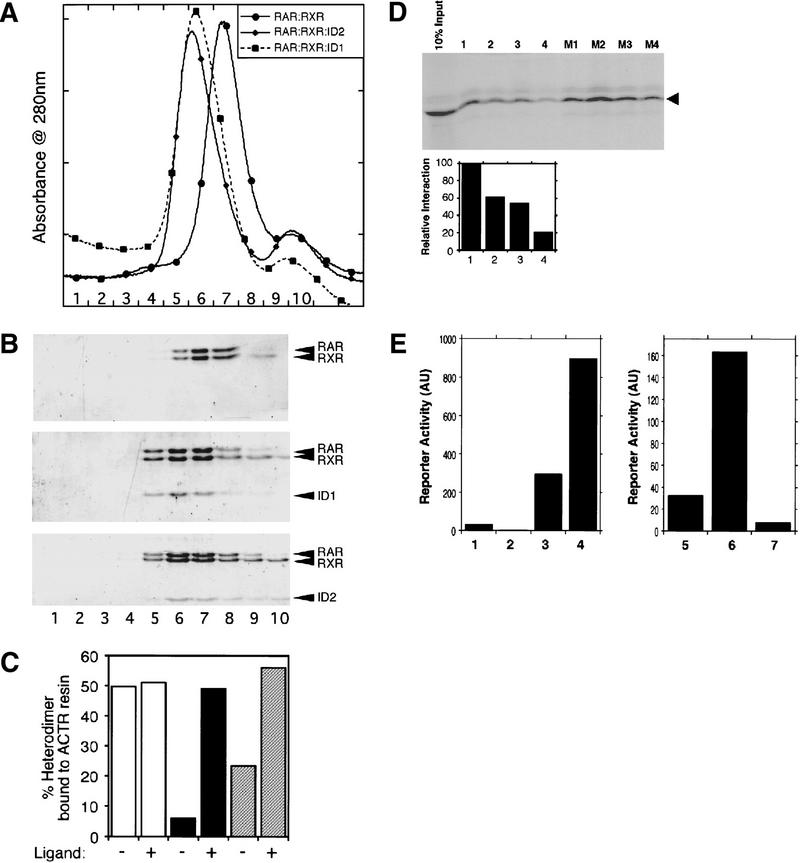
Figure 3.
In vitro analysis of the SMRT–ID : RAR–LBD : RXR–LBD complexes. (A) Gel filtration profiles of purified RAR–LBD : RXR–LBD heterodimers alone and with SMRT–ID1 [(SID1) amino acids 2202–2352], SMRT–ID2 [(SID2) 2098–2201]. (B) SDS-PAGE corresponding to A. Fraction numbers are indicated below both the profiles and gels. Note the relative elution volumes for the differently sized complexes. The heterodimer is smallest and therefore retarded most by the column. In each case, the elution profile and Coomassie staining in the gel are consistent with a 1:1:1 complex. This stoichiometry was supported by equilibrium centrifugation and light-scattering techniques. (C) Ligand facilitates the exchange of cofactors. The RAR agonist TTNPB increases the dissociation of SMRT–IDs from the RAR : RXR : SID1 (hatched bars) and RAR : RXR : SID2 (solid bars) complexes. This results in enhanced binding of RAR : RXR (open bars) to the GST–ACTR–ID resin. (D) In vitro competition assay with SMRT–ID2 (arrowhead) wild-type and mutant peptides. Radiolabeled ID2 was bound to GST–RXR–LBD : RAR–LBD heterodimers as described in Materials and Methods. This complex was competitively challenged by increasing the amount of wild-type synthetic peptide (lanes 1–4 using 0, 10, 30, and 100 μm peptide) or mutants 1–4 [M1–M4 at 30 μm (see Fig. 2)]. Quantitation of the wild-type competition assay is shown below. (E) (Left) SMRT minimal IDs can mediate ligand-independent activation of RAR. (Lane 1) Gal–DBD alone; (lane 2) Gal–RAR alone; (lane 3) Gal–RAR + VP–SMRT (ID1 + ID2); (lane 4) Gal–RAR + VP–SMRT (ID1 + ID2) + 10 nm TTNPB (RAR-specific agonist). (Right) Ligand-induced loss of activation of an RXR helix 12 mutant. (Lane 5) Gal–DBD alone; (lane 6) Gal–RXR mut (EK, EK) + VP–SMRT (ID1 + ID2); (lane 7) Gal–RXR mut (EK, EK) + VP–SMRT (ID1 + ID2) + 100 nm LG268 (RXR-specific agonist).